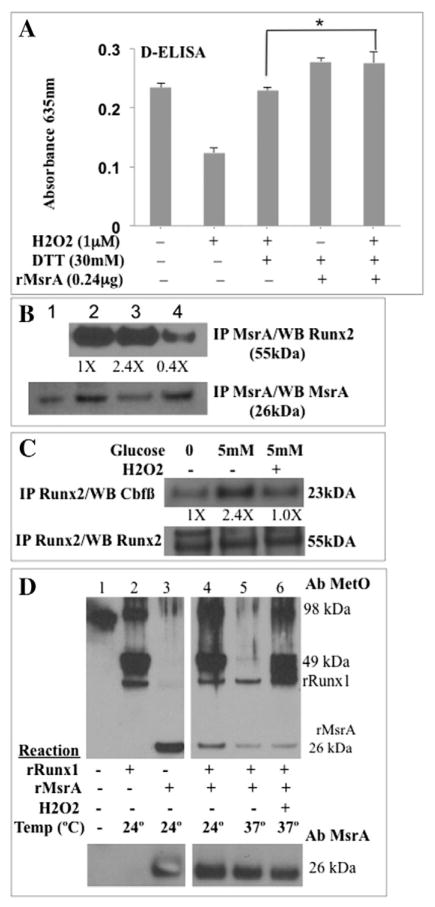
Fig. 6.
MsrA-regulated oxidation of RUNX methionine residues. (A) EC nuclear extracts were treated with the indicated compounds or with recombinant MsrA. Each treatment was performed in triplicate and RUNX2 DNA binding was monitored kinetically after the addition of HRP substrate (absorbance 635 nm) in a 96-well plate. Shown are results after 32 min (significant differences were noted between 10 and 32 min of reaction). p-Values relative to untreated (t-test): 1 μM H2O2 (p < 0.005); 1 μM H2O2 + 30 mM DTT (p > 0.05; NS); 30 mM DTT + MsrA or 1 μM H2O2 + 30 mM DTT + MsrA (p < 0.05). rMsrA increased RUNX2 DNA binding in the presence of H2O2 (*p < 0.05, ANOVA). (B) MsrA association with RUNX2 by co-immunoprecipitation (IP): representative gel from three determinations. ECs were treated in vivo with honokiol (10 μM) or H2O2 (100 μM). Nuclear extracts were isolated, immunoprecipitated with MsrA-specific antibody and immunoblotted with RUNX2 or MsrA-specific antibody. Recombinant MsrA control, lane 1; untreated cells, lane 2; cells + honokiol, lane 3; cells + H2O2, lane 4. Relative density of RUNX2 (normalized to MsrA) in each lane is indicated as fold changes. (C) Live cells were starved for 16 h (0 mM glucose) and treated with glucose (5 mM) or glucose + H2O2 (100 μM). RUNX2 antibody was used for immuneprecipitation of RUNX2-associated Cbfβ cofactor. Relative density of Cbfβ (normalized to Runx2) in each lane is indicated as fold changes. (D) RUNX1 (a surrogate for RUNX2) is an MsrA substrate. Recombinant proteins rRUNX1 or rMsrA were incubated individually or together at 24 °C or 37 °C for 30 min and resolved on SDS-PAGE. Western blot with specific antibody (Ab) detects Met-sulfoxide (MetO) or MsrA. Experiment was repeated with essentially similar results. Indicated are rRunx1 (49 kDa), rRunx1 dimers (98 kDa), and rMsrA (26 kDa).