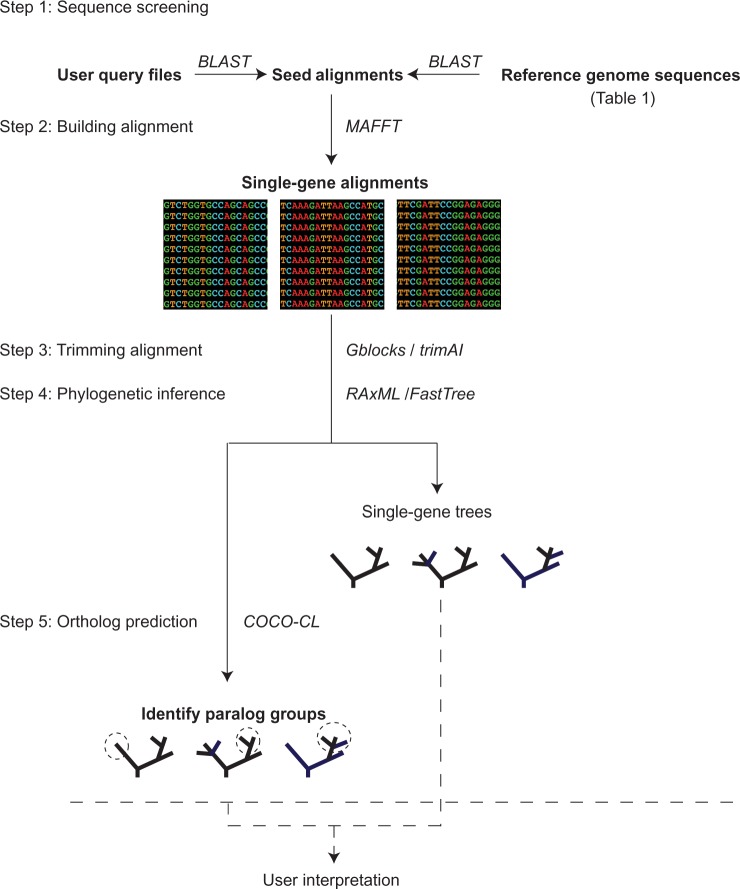
Figure 1.
Overview of steps in BIR pipeline. 1) The user provides a zipped file with the query sequences and another zipped file with the seed alignments. The sequence and alignments should be in FASTA format. Additionally, protein sequences from completely sequenced genomes (Table 1) can be added. Sequences from query files and selected reference genomes are added to the seed alignments with highest match using BLAST. 2) The modified seed alignments can be realigned using MAFFT. 3) Gblocks or trimAl can be used for removal of unambiguously aligned regions. 4) Phylogenetic trees can be inferred with FastTree or RaxML. 5) Paralog prediction is done by the COCO-CL program. Putative paralogs are marked in circles with a dashed line. The resulting phylogenetic trees can then further be assessed and interpreted using any tree-viewing software.