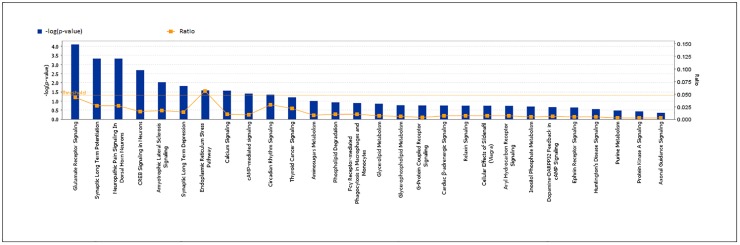
Fig 1. Canonical pathway analysis performed by IPA software including those genes tagged by SNPs with p-value <0.001 in our GWAs analysis.
The horizontal axis represents the pathways identified. The ratio (vertical axis, right) is calculated by the numbers of genes in a given pathway that meet cutoff criteria, divided by total numbers of genes that make up that pathway. The orange line stands for the threshold above which there are statistically significantly (by default P<0.05). The vertical axis (left) shows the −log of the p-value calculated based on Fisher’s exact test.