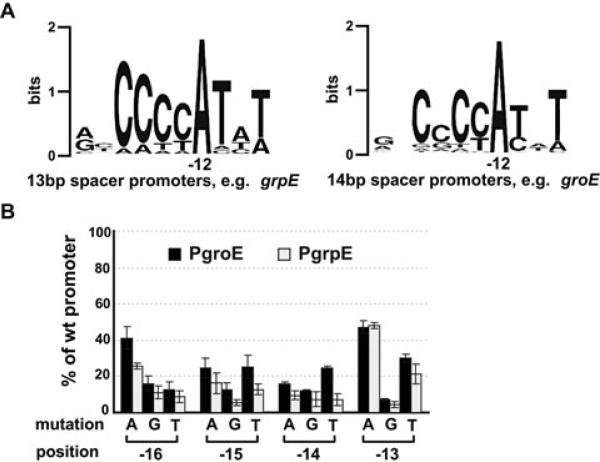
Fig. 2.
Sequence conservation in the −10 region of E. coli σ32 promoters and effects of single-base mutations at the −10 region tetra-C motif on activities of groE or grpE promoter.
A. Sequence logos (http://weblogo.berkeley.edu; Crooks et al., 2004) denoting sequence conservation within the −10 region of 13 bp spacer and 14 bp spacer promoters. Sequences of 16 σ32 promoters with a 13 bp spacer and 27 promoters with a 14 bp spacer (previously identified by Nonaka et al., 2006) were used to generate sequence logos of the −10 region.
B. Activity of each mutant groE or grpE promoter with wt σ32 is shown as a percentage of measured β-galactosidase activity of its respective wt promoter. Assay strains are as described in Fig. 1A. Promoter mutations and positions are shown on the x-axis. All values are averages of at least three independent experiments; error bars indicate one standard deviation; black bars, groE promoter activities; grey bars, grpE promoter activities.