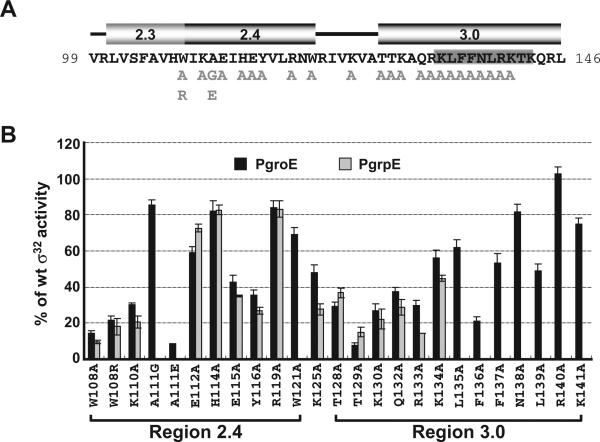
Fig. 3.
Effects of single amino acid substitutions in Regions 2.4 and 3.0 of σ32 on activity of the groE and grpE promoters.
A. Amino acid sequence of Regions 2.3, 2.4 and 3.0 of σ32. Numbers at each end of the sequence indicate amino acid positions. Single-amino acid substitutions of σ32 used in this study are shown in light grey below the main sequence. The location of the RpoH box is denoted as a grey box (Nakahigashi et al., 1995); positions of the αhelices in each region are shown above the sequence. These data are derived both from structural data indicating that these s subregions are virtually identical in σ70, a Group 1 σ, and σ28, a Group 3 σ (Campbell et al., 2002; Sorenson et al., 2004), and secondary structure predictions for σ32 that are completely consistent with these assignments (E. Campbell, data not shown).
B. Activity of the σ32 Regions 2.4 and 3.0 mutants at the wt groE and grpE promoters. β-galactosidase activity driven by each mutant σ32 from the wt groE or grpE promoters is shown as a percentage of that driven by wt σ32. Assay strains are as described in Fig. 1A. The different σ32 amino-acid substitutions and their locations are shown on the x-axis. Note the effects of several σ32 substitutions (W108R, A111G, A111E, W121A and from L135 to K141) were not measured at the grpE promoter. All values are averages of at least three independent experiments; error bars indicate one standard deviation; black bars, groE promoter activities; grey bars, grpE promoter activities.