Fig 2. Phylogenetic analysis of ZzPDS.
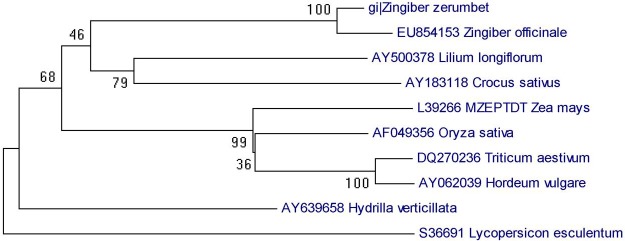
Phylogenetic analysis of Z. zerumbet phytoene desaturase (PDS) gene homologs was performed among EST sequences of Z. officinale (EY854183), Lilium longiflorum (AY 500373), Crocus sativus (AY 183118), Zea mays (L39266), Oryza sativa (AF 049356), Triticum aestivum (DQ 270236), Hordeum vulgare (AY 062039), Hydrilla verticillata (AY 639658), and Lycopersicon esculentum (S 36691). The phylogenetic tree was inferred using the Neighbor-joining method and tested by bootstrapping with 10000 replications. Bootstrap values >50% are shown at the nodes. The length of each branch represents the number of nucleotide substitutions per site.
