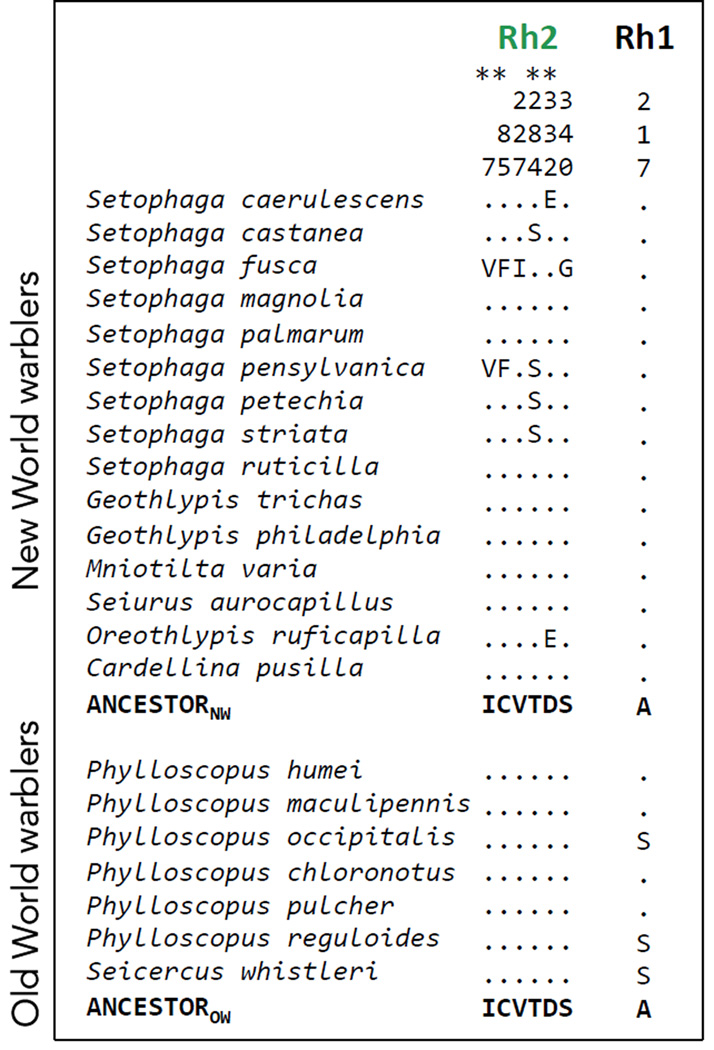
Figure 2.
Variable amino acid residues in the coding sequences of Rh2 and Rh1 opsins in the New World warblers and the Old World warblers. Numbers correspond to amino acid positions standardized by the bovine rhodopsin (Genbank M21606). The first 15 species are New World warblers and the following 7 are Old World warblers. All variable sites are shown relative to the inferred ancestor, which is the same for both clades, as obtained by Empirical Bayes methods (Yang 2005). Dots indicate the identity of the amino acids with the ancestor at each site, thus species that only have dots match the ancestral amino acid sequence at all sites. Residues marked with an asterisk (*) are inferred to evolve under positive selection by maximum likelihood methods. See Fig. S2 for comparisons across all Passerines.