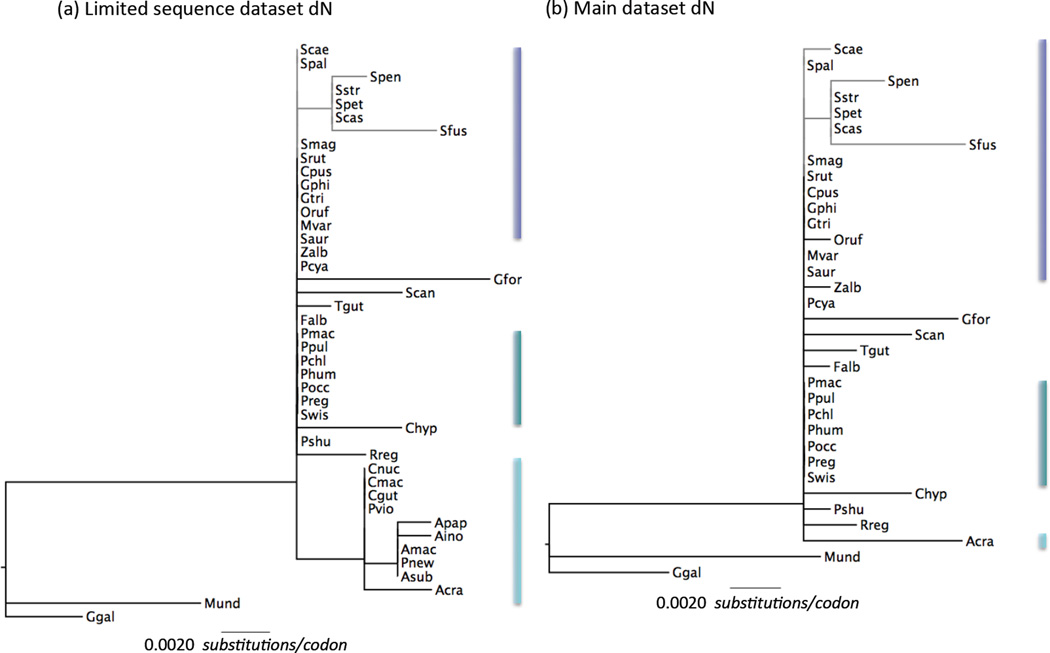
Figure 4.
Variation in Rh2non-synonymous rates of evolution (dN) across Passerines. Gene tree for Rh2 where branch lengths are scaled by dN rate (substitutions per codon) estimated along that branch by a free-ratio model as implemented in PAML (parameters specified in Table S8 for the best fitting codon model; Yang 2007). (a) the limited sequence dataset including bowerbirds for which incomplete coding sequences were used and (b) the full sequence dataset. Bars next to clades are colored following the scheme in Figs. 1 and 3 to highlight the position of each clade on the tree. Scale bar indicates value of dN and is the same in both trees. Species names are abbreviated as the first letter of the genus and the three first letters of the species (i.e. Setophaga castanea is Scas, see Figs. 1 and 2 for full species names).