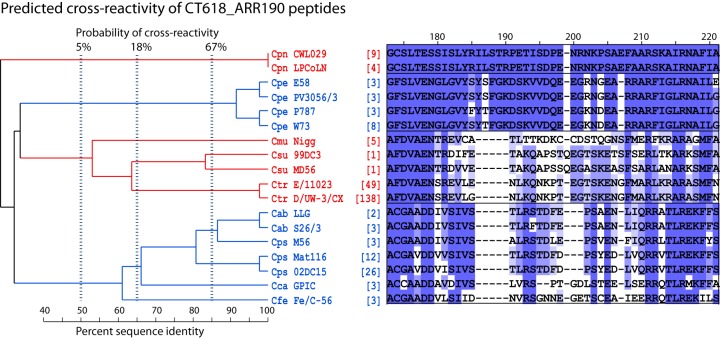
FIG 3.
Evolutionary relationship and predicted cross-reactivity among CT618_ARR190 sequences. Numbers at the top of the subregion alignment indicate approximate residue numbers of the Ctr_CT618 protein in the alignment of the overall region (Fig. 1). Corresponding residues in mismatched sequences are not always identical to the overall ARR alignment because of additional sequences, different sequence lengths, gap insertions, and higher gap insertion penalty in the subregion alignments. Numbers in brackets indicate the frequency of the strain-specific sequence available in the NCBI protein database. For clarity, deep clades (sequences of typically <50% amino acid identity) are indicated in alternating color, with the sequences separated into boxes. Vertical dashed lines indicate the calculated probabilities of antibody cross-reactivity among peptides with a single common ancestor at these thresholds. This ARR shows high evolutionary divergence, allowing for robust differentiation of a chlamydial species (Cpn or Cpe) or a group of closely related chlamydial species (clade Cmu, Csu, and Ctr and clade Cab, Cps, Cca, and Cfe). Cps species/strain-specific peptides are evolutionarily closest to Cab, followed by the Cca and Cfe species/strain. Therefore, Cps peptides have the highest probability of cross-reactivity with Cab. Cpn or Cpe peptide sequences are evolutionarily well separated, and these peptides have a very low probability of serological cross-reactivity with the remaining eight chlamydial species.