Abstract
DNA cytosine methylationis an important epigenetic process that is correlated with transgene silencing, transposon suppression, and gene imprinting. In plants, small interfering RNAs (siRNAs) can trigger DNA methylation at loci containing their homolog sequences through a process called RNA-directed DNA methylation (RdDM). In canonical RdDM, 24 nucleotide (nt) siRNAs (ra-siRNAs) will be loaded into their effector protein called ARGONAUTE 4 (AGO4) and subsequently targeted to RdDM loci through base-pairing with the non-coding transcripts produced by DNA-directed RNA Polymerase V. Then, the AGO4-ra-siRNA will recruit the DNA methyltransferase to catalyze de novo DNA methylation. Recent studies also identified non-canonical RdDM pathways that involve microRNAs or 21 nt siRNAs. These RdDM pathways are biologically important since they control responses biotic and abiotic stresses, maintain genome stability and regulate development. Here, we summarize recent pro-gresses of mechanisms governing canonical and non-canonical RdDM pathways.
Keywords: Plants, siRNA, ARGONAUTE, DNA methylation
INTRODUCTION
DNA methylation is an important modification of chromatin. It can cause inheritable alterations of genetic information and often correlates with the repression of gene expression [1-3]. In mammals, the majority of DNA methylation occurs at the symmetric CG context and ~ 70-80% of CG of the whole genome are methylated [3]. In plants, over 50% of the genome, including centromeric region and repetitive sequences, is methylated [1-3]. Cytosine methylation in plants occurs in three sequence contexts: CG, CHG, and CHH, where H represents any nucleotide other than guanine [1-3]. In mammals, DNA methylation is established by DNA methyltransferase 3 (DNMT3), the de novo DNA methyltransferase, which involves the interaction between DNMT3-LIKE protein and the unmethylated Histone 4 tail [3]. The DNA methylation of mammals is maintained by DNMT1, the DNA methyltransferase acting on hemi-methylated DNAs after replication [3]. In plants, DNA methylation can be established through a process called RNA-directed DNA methylation (RdDM), by which small interference RNAs (siRNAs) recruit the DNA methyl transferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to target loci containing their homolog sequences and catalyze de novo cytosine methylation at all sequence contents [4-6]. As a conserved de novo DNA methylation mechanism, RdDM is involved in diverse epigenetic processes such as transposon suppression and gene imprinting [7-11]. During DNA replication, the symmetric CHG and CG methylationcan be maintained by the DNA methyltranferase MET1 and CMT3, respectively, which recognize hemi-methylated DNA strand through mechanisms other than RdDM and addmethyl groups to the de novo synthesized DNA strand [12-15]. In contrast, the maintenance of asymmetric CHH methylation still needs RdDM to re-establish since both daughter strands do not contain cytosine methylation [15]. Recent studies from the model plant Arabidopsis thaliana have greatly increased our understanding of the RdDM process. Many components critical for RdDM have been identified by genetic and proteomic approaches. Studies on these genes have established the framework of RdDM. We here review recent progresses related to RdDM.
24 nt siRNA-TRIGGERED DNA METHYLATION
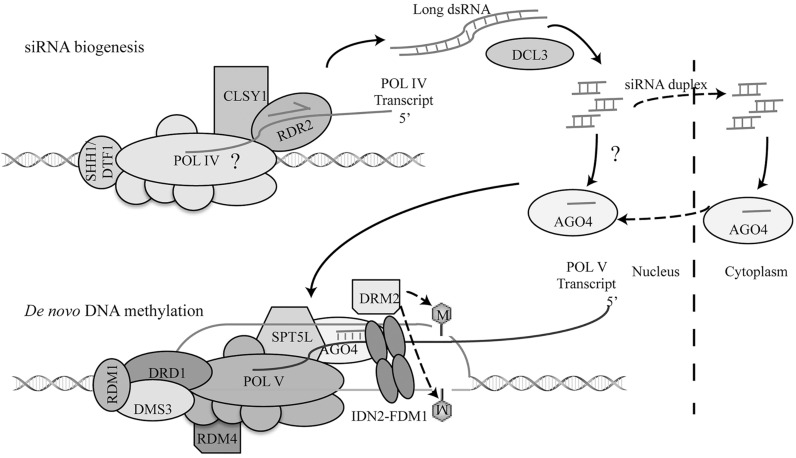
The process of RdDM starts with the biogenesis of ~ 24 nucleotide (nt) siRNAs(ra-siRNAs) from RdDM target loci. In this process, the plant specific DNA-directed RNA Polymerase IV (Pol IV) is thought to produce single-stranded RNAs (ssRNAs) from RdDM target loci, which will be converted to double-stranded RNAs (dsRNAs) by the RNA-dependent RNA Polymerase 2 (RDR2) (Fig. 1) [16-19]. The resulting dsRNAs will be processed into ra-siRNAs by the RNAse III enzyme DICER-LIKE 3 (DCL3) [19]. Following production, ra-siRNAs are methylated by HEN1, which protects them from 3’ untemplated uridine addition (Uridylation) and degradation [20, 21], and, then are loaded into their effector protein named ARGONAUTE 4 (AGO4) in cytoplasm [22]. After imported into nucleus, the AGO4–ra-siRNA complex recruits DRM2 to catalyze de novo DNA methylation through targeting the nascent scaffold transcripts generated by the plant specific DNA-directed RNA Polymerase V (Pol V) (Fig. 1) [23, 24]. Besides these major players, RdDM also requires the assistance from many accessory proteins (Fig. 1).
Fig. (1).
The framework of canonical RNA-induced DNA methylation. With the assistance of SHH1, Pol IV is recruited to chromatin to generate single-stranded RNAs. RDR2 then converts ssRNAs to dsRNAs, which are processed into 24 nt ra-siRNAs by DCL3. The optimal activities of RDR2 and Pol IV require CLSY. After assembly in the cytoplasm, AGO4-ra-siRNA complexes are imported into nucleus, recruited to chromatin through AGO4-Pol V interaction and base pairing between ra-siRNAs and transcripts of Pol V. The DDR complex composed of RDM1, DMS3 and DRD1 is required for the Pol V-chromatin interaction and the activity of Pol V. AGO4-rasiRNA complex and base-pairing between ra-siRNA and transcripts of Pol V may recruit DRM2 to methylate DNA, which requires SPTL5 and the IDN2-FDM1 complex.
DICER-LIKE Proteins Involved in RdDM
Dicer proteins are members of ribonuclease III (RNaseIII) family that process dsRNAs to release ~21-24 bp RNA duplexes with a 5’ phosphate and a 2nt 3’ overhang at each strand [25]. Dicer proteins contain several domains: DEAD box, helicase-C, DUF283, PAZ, two RNase III domains, and dsRBD [25]. Among these protein domains, two RNaseIII domains forman intermolecular dimer, each of which hydrolyzes one strand of dsRNAs [26]. The PAZ domain is critical for substrate recognition since it is connected with RNaseIII domain by a long α helix and binds the 3’ terminal nucleotide of a dsRNA with a 2nt 3’ overhang, and therefore, [26]. It is proposed that the distance between PAZ and the catalytic site of RNase III domain determines the length of cleavage products of Dicer [26].
Plants encode several DICER-LIKE proteins (DCL) to meet the requirement of multiple small RNA pathways [25]. Four Dicer-like genes exist in Arabidopsis, DCL1-DCL4 [25]. Among of them, DCL3 is the major enzyme responsible for the production of 24 nt ra-siRNAs from long double-stranded RNAs (dsRNAs) [19]. DCL3 requires protein factors for its optimal activity. The RNA binding protein TOUGH (TGH) interacts with DCL3 and is required for efficient dsRNA processing, suggesting that TGH may act as a partner to facilitate DCL3 function [27]. In addition, two RNA-binding proteins MOS4 and DAWDLE (DDL) and the DNA-binding protein CDC5 are also required for ra-siRNA accumulation [28-30]. DDL and CDC5 interact with DCL1, a homolog of DCL3 [28]. By analog, they may also interact with DCL3 to promote its activity [28]. Although DCL3 is essential for the production of 24 nt ra-siRNAs, lack of DCL3 only moderately affects DNA methylation, suggesting other DCLs may have overlapping functions with DCL3 in RdDM [31]. In fact, at some loci, DNA methylation can be completely eliminated in the dcl2 dcl3 dcl4 triple mutant [31]. In addition, DCL3 can also generate 24 nt siRNAs from transcripts of Geminiviruses that have single-stranded DNA genomes. The resulting 24 nt viral-derived siRNAs are loaded into AGO4 and direct viral DNA methylation, which helps the host to defense the virus. However, this process does not involve RDR2 [32].
The Plant Specific Pol IV and Pol Vare Essential for RdDM in Arabidopsis
Plants contain two specific DNA-dependent RNA Polymerases, named as DNA-dependent RNA Polymerases IV (Pol IV) and Pol V (Pol V) [33]. Pol IV and Pol V are conserved among different plant species, such as Arabidopsis, rice, and maize. Pol IV and Pol V are large protein complexes, with molecular mass close to 1 Mega dalton [33]. They are Pol II-derived Polymerase, as they contain many identical and paralogous subunits of Pol II [33-35]. Pol IV and Pol V share eight identical subunits, including the second largest subunit NRPD2/NRPE2 [33-35]. They also have four paralogous subunits, including the largest subunits NRPD1of Pol IV and NRPE1 of Pol V [33-35]. Although paralogous subunits have some highly conserved features, they have considerable differences. For instance, the N-terminal regions of NRPD1, NRPE1, and NRPB1 (largest subunit of Pol II) are highly conserved and all the three Polymerases contain evolutionary conserved regions A to H [18]. However, their C-termini show variations, which are proposed to cause differences in Polymerase activities of Pol II, IVand V [18]. Different from the C-terminal domain (CTD) of NRPB1, the C-terminal of NRPD1 shares similarity with the C-terminal half of a nuclear-encoded protein named DEFECTIVE CHLOROPLAST AND LEAVES (DCL), which regulates rRNA processing in chloroplasts [18]. Compared with NRPB1 and NRPD1, NRPE1 shows additional features in its C-terminal, including a long CTD that extends beyond the DCL-like motif and multiple potential phosphorylation sites in a highly hydrophilic domain composed by ten complete repeats of a 16-amino-acid consensus sequence [18]. Protein sequence differences among paralogous subunits of Pol II, IV and V often occur in the key positions relative to the template channel and RNA exit paths, which may cause the functional divergence among Pol II, IV and V [33-35].
Loss-of-function mutations in NRPD1 and NRPE1 abolish DNA methylation at RdDM target loci, establishing the essential role of Pol IV and Pol V in RdDM [16-18, 36]. However, nrpd1 and nrpe1 have different impacts on RdDM. The majority of siRNAs (>94%) are dependent on NRPD1 for accumulation, demonstrating that Pol IV acts in siRNA production [7, 8]. In contrast, many of the Pol IV-dependent siRNAs do not require Pol V for their accumulation although Pol V is required for the methylation of these siRNA-generating loci [7]. This result suggests that Pol V have roles other than siRNA production in RdDM [7]. The study on low-abundance intergenic noncoding (IGN) transcripts that are produced from flanking regions of RdDM loci in Arabidopsis revealed the role of NRPE1 in DNA methylation [37]. NRPE1 associates with the IGN region and is required for the accumulation of IGN transcripts, suggesting Pol V may be responsible for the transcription of IGN RNAs [37]. The Pol V-dependent transcripts from IGN regions are independent ofPol IV, DCL3 or RDR2 [37]. In nrpe1, the association of AGO4 with target DNA loci is disrupted, suggesting these Pol V-dependent transcripts may act as scaffolds to recruit the downstream silencing machinery, such as AGO4 [24].
Besides NRPD1 and NRPE1, NRPD2/ NRPE2, the second largest subunit of Pol IV and Pol V and several smaller subunits of Pol IV and Pol V, are also studied by mutation analysis. Without NRPD2/NRPE2, both ra-siRNAs and DNA methylation are almost undetectable, revealing that NRPD2/NRPE2 is essential for the function of both Pol IV and Pol V [16-18, 36]. The other well-studied subunit required for RdDM is NRPD4/NRPE4, which is also a common subunit for Pol IV and Pol V. NRPD4/NRPE4 shares sequence similarity with NRPB4, a subunit of Pol II, but has different functions. NRPD4/NRPE4 is physically associated with NRPD1 and NRPE1 [38]. Loss-of-function mutations in NRPD4/NRPE4 mutants reduce DNA methylation levels and siRNA accumulation, illustrating that NRPD4/NRPE4 is involved in RdDM [38]. In addition, NRPE5 and NRPE 9bwere found to function exclusively in PolV [39]. However, mutation analysis did not reveal the function of other subunits of Pol IV and Pol V in RdDM, suggesting that they are redundant or that they are non-essential for the function of Pol IV and Pol V in RdDM [33-35].
Protein Factors Required forPol IV and Pol V Function
In eukaryotes, Pol II requires transcription factors for its activity. Like Pol II, Pol IV and Pol V also require protein partners. Pol II has been shown to assist the recruitment of Pol IV and Pol V to chromatin at some loci presumably through its transcription activity [40]. CLSY1, a putative chromatin-remodeling factor, is required for the production of 24 nt ra-siRNAs and the spreading of transcriptional gene silencing (TGS) signals [41]. In clsy1, NRPD1 becomes diffused localization pattern in nucleoplasm from the discrete loci that are shown in wild-type plants. This result suggests that CLSY1 is required for proper Pol IV localization [41]. The recruitment of Pol IV to chromatin requires SAWADEE HOMEODOMAIN HOMOLOGUE 1 (SHH1), which interacts with Pol IV and binds to H3K9me and unmethylated H3K4 through its unique tandem Tudor-like fold [42-45].
SUPPRESSOR OF TY INSERTION5-LIKE (SPT5L, also known as KTF1), a homolog of the yeast transcription elongation factor Spt5, was identified as a potential transcription factor associated with NRPE1 [35, 46, 47]. The chromatin association of Pol V is independent of SPT5L [48]. However, SPT5L requires Pol V for its proper chromatin association at RdDM target loci,demonstraing that SPT5L acts downstream of Pol V, which is consistent with the transcription elongation function of its yeast homolog [48]. Besides SPT5L, a homolog of yeast transcription factor IWR1 termed RDM4/DMS4 is shown to affect the accumulation of 24 nt siRNAs [49, 50]. rdm4/dms4 loss-of-function mutants show significant reduction in Pol V-dependent transcripts, suggesting that Pol V requires the assistance of RDM4/ DMS4 for its proper function [49, 50]. Consistently, RDM4/DMS4 physically interacts with NRPE1, which further supports the role of RDM4/DMS4 in POL V transcription [49]. The activity of Pol V also requires a DDR complex composed of DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), a putative chromatin-remodeling factor with SNF2 domain, DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), a protein with a domain that is similar to the hinge region of structural maintenance of chromosome proteins (SMC), and RDM1, a plant specific small protein [51-53]. The DDR complex is required for the recruitment of Pol V to chromatin and therefore the production of Pol V-dependent transcripts [52, 53]. In addition, RDM1 also serves as a linker between AGO4 and DRM2 [52, 53]. Three members of the SU(VAR)3-9 histone methyltransferase family, SUVH2, SUVH9 and SUVR2, can bind methylated DNA through their SET and RING-associated domain, and, therefore, facilitate the Pol V-chromatin interaction at some loci with existed DNA methylation [54-57].
RNA-dependent RNA Polymerases involved in RdDM
RNA-dependent RNA Polymerase (RDR) can use ssRNA molecule as a template to synthesize dsRNA [58, 59]. These proteins have been identified in plants, fungi, C. elegans, but not in mice or human. Eukaryotic RDRs can be classified into three major types: RDRα, RDRβ, and RDRγ. RDRα exists in both plants and lower animals [58, 59]. RDRβ is specific in lower animals, while RDRγ is specific in plants. There are six RDRs in Arabidopsis, RDR1-6 [58, 59]. Among six RDRs, RDR1, 2, and 6 share the C-terminal canonical catalytic DLDGD motif of eukaryotic RDRs [58, 59]. Initially, the three RDRs were thought to be involved in plant anti-virus mechanism, such as posttranscriptional gene silencing (PTGS). However, they have evolved unique biological functions. RDR2 is the only one participating in 24 nt siRNA-mediated DNA methylation [19]. In the null rdr2 mutants, miRNA and ta-siRNA production is unaffected, suggesting that RDR2 functions specifically in RdDM pathway [60]. Consistent with the loss of ra-siRNAs production, DNA methylation levels in rdr2 mutants are reduced significantly [19]. RDR2 co-localizes with NRPE1, DCL3, and AGO4 at discrete spots called Cajal bodies, the conserved sites for the maturation of ribonucleoprotein complexesin nucleolus, suggesting the presence of the nucleolar processing center for ra-siRNA production [61, 62]. The facts that 98.5% of Pol IV-dependent siRNAs are lost in rdr2 mutant and that Pol IV and RDR2 are physically associated in vivo suggest that RDR2 may function together with Pol IV to synthesize dsRNAs [43]. Consistent with this notion, in vitro biochemical studies show that RDR2’s Polymerase activity depends on Pol IV [63]. In the absence of Pol IV, RDR2 dose not synthesize RNA fragments using DNA-RNA bipartite templates [63].
Two biochemical activities of RDR2 have been proposed based on the studies of a Neurospora RDR gene termed QUELLING DEFECTIVE1 (QDE1), which acts in RNA silencing and DNA repair pathways [63]. QDE1 has at least two activities. It can synthesize a serious discontinuous RNA along the template RNA without primers [64]. It can also use the free 3’ terminal of a RNA template as a primer to synthesize full-length complementary RNA [64]. According to the dual activities of QDE1, RDR2 may move together with Pol IV along DNA and synthesize a series of discontinuous second strands from the internal of Pol IV-dependent transcripts before the termination of Pol IV transcription, which is analogous to lagging-strand Okazaki fragment generated during DNA duplication [63]. Alternatively, RDR2 may use complete transcripts of Pol IV as templates to generate the full-length fragment [63].
The ARGONAUTE (AGO) Proteins Involved in RdDM
ARGONAUTE (AGO) proteins are the effector proteins in small RNA-induced gene silencing pathways [65]. AGO usually contains four major domains: N-terminal domain, PAZ, MID and PIWI domains. The PAZ domain binds to the 3’ end of small RNA and the MID domain binds to the 5’ end of small RNA [65]. The PIWI domain shows similarity to ribonuclease-H enzyme with conserved Asp-Asp-Asp/Glu/His/Lys motif and is responsible for the cleavage of target mRNAs. However, not all AGOs have such slicer activities [65].
There are ten AGO proteins (AGO1-AGO10) in Arabidopsis. Among of them,AGO4, AGO6 and AGO9 act in RdDM [65, 66]. The function of AGO4 in RdDM has been extensively studied. AGO4 binds RdDM loci and plays essential role in DNA methylation and ra-siRNA amplification, which is independent of its slicer activity [24, 67]. AGO4 co-localizes either with NRPE1 in Cajal bodies, which are dynamic compartments for siRNA processing, or with NRPE1, NRPE2 and DRM2 at a separate discrete nuclear body termed as the AGO4-NRPE1 (AB) body, which is a potential active site for RdDM [62]. Further studies show that AGO4 physically interacts with NRPE1, the largest subunit of Pol V, through the GW/WG repeats in the CTD region of NRPE1 [23]. AGO4 associates with the Pol V-dependent transcripts through base pairing between ra-siRNAs and Pol V-dependent transcripts. In nrpe1, the AGO4-chromatin association is disrupted, suggesting that the Pol V-AGO4 interaction and the association of AGO4-siRNA with Pol V-dependent transcripts may recruit AGO4 to the RdDM loci [24]. The recruitment of AGO4 to Pol V-dependent transcripts needs the assistance from SPT5L/KTF1, which interacts with AGO4 through its GW/WG repeats motif [46, 48]. SPT5L also bind POL V-dependent transcripts, indicating that SPT5L may function as an adaptor for AGO4 and POL V-dependent transcripts [48].
RdDM was thought to be solely nuclear process because both biogenesis and functioning of ra-siRNAs take place in nucleus. However, a recent study shows that the levels of individual 24 nt ra-siRNAs is ~ ten times higher in cytoplasm than those in in the nucleus [67]. The majority of cytoplasmic ra-siRNAs exist as a duplex form, while 24 nt siRNAs in nucleus are single-stranded [67]. Furthermore, a small fraction of AGO4 associates with only single-stranded cytoplasmic 24 nt siRNAsin the cytoplasm [67]. These results reveal that the loading of siRNAs into AGO4 may occur in cytoplasm and the AGO4-siRNA complexesmay need to be imported into nucleus for their functions [67].
The SGS3-LIKE Proteins Involved in RdDM
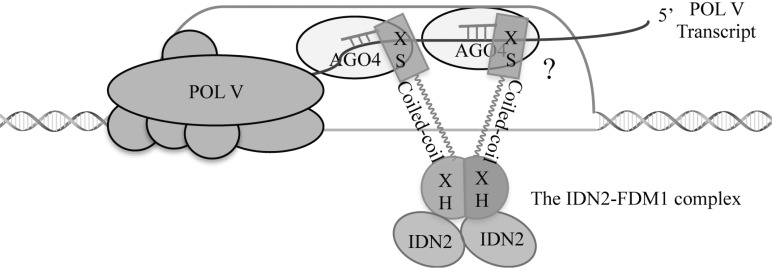
RdDM also requires INVOLVED INDE NOVO 2 (IDN2, also called RDM12), FACTOR1 of DNA METHYLATION 1 (FDM1; also called IDN2 PARALOG 1; IDP1 or IDN2-LIKE 2; IDL1), FDM2 (IDP2, IDL2), FDM3, FDM4 and FDM5 [68-73]. These proteins are members of plant specific SGS3-LIKE protein. SGS3, the founding member, is a key player in transgene-induced post-transcriptional gene silencing (PTGS) [74]. IDN2 interacts with the XH domain of either FDM1 or FDM2 to form a tetramer containing 2 copies of each protein, which is required for RdDM [68, 70, 73]. Both IDN2 and FDM1 bind dsRNAs with 5’ overhangs, a structure resembling dsRNAs formed by base-pairing between siRNAs and Pol V-transcripts in vitro (Fig. 2) [69, 70]. In addition, FDM1 can bind unmethylated but not methylated DNA through its coil-coil domain, which is independent ofits RNA binding ability (Fig. 21) [70]. These results suggest that the IDN2-FDM1/FDM2 complex may be recruited to the chromatin by the ra-siRNA-Pol V transcript duplex, and then bind the unmethylated DNA to initiate but not reinforce DNA methylation (Fig. 2) [69, 70]. Consistent with this hypothesis, RNA can mediate the AGO4-FDM1 interaction and IDN2 binds Pol V-transcript in a siRNA- and AGO4-dependent manner, which is required for the association of DRM2 with Pol V-dependent transcripts and de novo DNA methylation [70, 75]. IDN2 also interacts with the SWI/SNF chromatin-remodeling complex, which alters the nucleosome position and facilitates RdDM [76]. Although the function of FDM3-5 in RdDM has been indicated, their functional mechanisms remain to be identified.
Fig. (2).
Proposed functional model for the IDN-FDM1 complex. The IND2 and FDM1 proteins form a tetramer containing 2 copies of each protein. The IDN2-FDM1 interaction is mediated by their XH domains. The IDN2-FDM1 complex binds dsRNAs formed by base pairing between ra-siRNAs and Pol V-dependent transcripts through their XS domains. FDM1 can bind unmethylated DNA through its coil-coil domain, which may facilitate DRM2 to recognize target loci.
METHYLTRANSFERASES INVOLVED IN DE NOVO METHYLATION BY RDDM
Three methyltransferases have been identified that are involved in plant DNA methylation: DRM2, DNA METHYLTRANSFERASE 1 (MET1), and CHROMOMETHYLASE 3 (CMT3) [12-14, 77]. MET1 is the plant homolog of DNMT1, which is the methyltransferase responsible for maintaining DNA methylation patterns during cell division in mammals [12]. DRM2 also has a mammalian homolog termed DNMT3, the de novo methyltransferase setting up DNA methylation patterns in the early stage of development [77]. However, CMT3 is a plant specific methyltransferase without any mammalian homolog [13, 14]. DRM2, MET1 and CMT3 have different functions in the establishment and maintenance of DNA methylation during cell division [12-14, 77]. The establishment of DNA methylation is mainly catalyzed by DRM2 via de novo methylation [77]. DRM proteins form a homodimer for their catalytic activity [6]. In addition, DRM2 associates with the AGO4-ra-siRNA complex and shows biased methylation on the template DNA strand for Pol V [6]. This result suggests that the recruitment of DRM2 to target loci involves AGO4-ra-siRNA and base-pairing between siRNAs and Pol V-dependent transcripts [6]. DRM2, MET1, and CMT3 all participate in DNA methylation maintenance. However they act in different sequence contexts:CG methylation by MET1, CHG methylation by CMT3, and CHH methylation by DRM2 [12-14, 77].
The Role of Histone Modification and Chromatin Structure in RdDM
Histon H3 Lys 9 (H3K9) methylation, which overlaps with most of RdDM target [78], has been shown to play critical roles in the reinforcement of DNA methylation. Lack of KRYPTONITE (SUV4), SUV5 and SUV6, the methyltransferase specific to H3K9, reduces the methylation at CHG site [79-81]. These Histone methyltransferases and CMT3 bind methylated DNA and methylated H3K9, respectively, and thus form a self-reinforcing loop for both H3K9 methylation and DNA methylation [54, 79]. In addition, the Jumonji C-type histone demethylases JUMONJI 14 (JMJ14) LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 (LDL1) and LDL2, which remove H3K4 methylation, UBIQUITIN-SPECIFIC PROTEASE 26 (UBP26), which is responsible for deubiquitylation of Histone H2,and HISTONE DEACETYLASE 6 (HDA6) are also required for RdDM, suggesting that the elimination of active markers of histone is an essential step in RdDM [82-87]. Furthermore, loss-of-function mutations in two microrchidia (MORC)-type ATPases, MORC2 and MORC6, which may be required for higher-order chromatin condensation, cause moderate loss of DNA methylation, revealing that the chromatin condensation may have roles in the reinforcement of DNA methylation [88, 89].
NON-CANONICAL RdDM MECHANISMS
In plants, DNA methylation can also be induced by non-canonical RdDM mechanisms, such as microRNA (miRNA)-, ta-siRNA- and 21 nt siRNA-induced DNA methylation. These processes often need the protein factors that are involved in PTGS besides some components essential for canonical RdDM.
In Arabidopsis, RDR6, a homolog of RDR2, has been shown to convert some RNAs produced by Pol II to dsRNAs, which can be processed by DCL2 or DCL4 into 21 or 22 nt siRNAs. These siRNAs often associate with AGO1 to direct target cleavage, and thus silence gene expression at post-transcriptional levels. A recent study shows that some RDR6-depedent siRNAs can initiate de novo DNA methylation at some transposon loci where the siRNAs are originated [90]. This process depends on AGO2, Pol-V transcripts and DRM2 [90]. The resulting DNA methylation triggers the production of 24 nt siRNAs to initiate the canonical RdDM pathway, which reinforces DNA methylation at the target transposon loci [90]. A research studying de novo silencing of a newly integrated retrotransposon shows that RDR6-dependent dsRNAs can be processed to 24 nt ra-siRNAs to trigger canonical RdDM when their levels are saturated for DCL2 and DCL4 [91]. This result indicates the variations of RDR6-dependent RdDM in plants. In addition, analyses of NERD, which is a GW repeat- and PHD finger-containing protein and binds Histone H3 and AGO2, reveal the presence of additional RdDM pathway [92]. The NERD-dependent RdDM requires Pol IV, Pol V, 21 nt siRNAs, AGO2, RDR6, and several proteins required for RDR6 pathway including SGS3, SILENCING DEFECTIVE 3 (SDE3; a RNA helicase) and SDE5 (a putative mRNA export factor) [92, 93]. NERD-dependent and RDR6-dependent RdDM may have overlapped mechanism since many protein factors function in both pathways.
microRNAs (miRNAs) are another class of riboregulator that represses gene expression. After processed by DCL1 from Pol II-dependent stem-loop transcripts (pri-miRNAs), most miRNAs bind AGO1 to direct cleavage and/or translational inhibition of target mRNAs. In rice, some pri-miRNAs can be processed by DCL3 to produce 24 nt miRNAs [94]. Unlike DCL1-dependent 24 nt miRNAs, these DCL3-dependent miRNAs are sorted into AGO4 to trigger DNA methylation at miRNA encoding loci or target loci likely using a canonical RdDM mechanism [94]. In the moss Physcomitrella patens, the ratio between miRNAs and their targets activates the miRNA-triggered DNA methylation. Ta-siRNA is a class of siRNA that triggers the degradation of target RNA. Ta-siRNAs are processed by DCL4 from dsRNAs, which are converted by RDR6 from miRNA-cleavage products [95]. However, some RDR6-dependent dsRNAs can be processed by DCL1 to produce siRNAs, which are loaded into AGO4 or AGO6 to trigger DNA methylation using canonical RdDM mechanism [95].
Prospective
Although studies have established the framework of RdDM, many questions need to be addressed. Pol IV is thought to transcribe ssRNA from DNA templates. However, the in vivo products of Pol IV remain to be identified. Pol IV and Pol V are similar to Pol II. Additional questions include how Pol IV and Pol V-dependent transcriptions are initiated and whether the Pol IV and Pol V activities are subjected to regulation. The recruitment of Pol IV to RdDM loci still needs additional factors since SHH1 only is responsible for a subset of DNA loci [44]. In addition, how the RdDM target loci are distinguished from non-RdDM DNA loci to initiate siRNA production is not clear yet. Although some proteins are required for RdDM, their functional roles remain to be defined. The recruitment of Pol IV and Pol V appears to require the existence of histone marker. How these histone markers are established needs further investigation. Although Pol II has roles in the recruitment of Pol IV and Pol V to chromatin, its functional mechanism is not known yet. In maize, the reduction of RdDM causes pleotropic development defects [1]. This is different with the observation in Arabidopsis, suggesting that RdDM functions maybe varied in different plant species and need to be further explored. The involvement of RdDM in plant adaption to environmental alterations as wellas abiotic stresses has been identified [1]. Further studies need to elucidate how RdDM is controlled in response to abiotic and biotic stimuli and whether the resulting alteration of DNA methylation can be inherited subsequently to benefit the progeny.
ACKNOWLEDGEMENTS
We thank Dr. Shuxin Zhang from University of Nebraska, Lincoln for critical reading of the manuscript. This work was supported by a National Science Foundation Grant MCB-1121193 (to B.Y)
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
References
- 1.Matzke M.A., Mosher R.A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 2.Castel S.E., Martienssen R.A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14(2):100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303(5662):1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 5.Mette M.F., Aufsatz W., van der Winden J., Matzke M.A., Matzke A.J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19(19):5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157(5):1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105(8):3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA. 2007;104(11):4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 10.Gehring M., Bubb K.L., Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324(5933):1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibarra C.A., Feng X., Schoft V.K., Hsieh T.F., Uzawa R., Rodrigues J.A., Zemach A., Chumak N., Machlicova A., Nishimura T., Rojas D., Fischer R.L., Tamaru H., Zilberman D. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vongs A., Kakutani T., Martienssen R.A., Richards E.J. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 13.Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15(14):1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292(5524):2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 15.Chan S.W., Henderson I.R., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6(5):351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 16.Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308(5718):118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 17.Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37(7):761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 18.Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120(5):613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2(5):E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15(16):1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G., Chen X., Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 2012;22(8):695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 23.El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21(20):2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41(5):630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010;20(1):49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 26.Hammond S.M. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579(26):5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 27.Ren G., Xie M., Dou Y., Zhang S., Zhang C., Yu B. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109(31):12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Xie M., Ren G., Yu B. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. USA. 2013;110(43):17588–17593. doi: 10.1073/pnas.1310644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C.J., Zhou J.X., Liu J., Ma Z.Y., Zhang S.W., Dou K., Huang H.W., Cai T., Liu R., Zhu J.K., He X.J. The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 2013;32(8):1128–1140. doi: 10.1038/emboj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38(6):721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 32.Raja P., Sanville B.C., Buchmann R.C., Bisaro D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008;82(18):8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag J.R., Pikaard C.S. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12(8):483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 34.Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolić L., Pikaard C.S. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell. 2009;33(2):192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L., Jones A.M., Searle I., Patel K., Vogler H., Hubner N.C., Baulcombe D.C. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 2009;16(1):91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 36.Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19(17):2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierzbicki A.T., Haag J.R., Pikaard C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135(4):635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.S., Zhu J.K. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23(3):318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eun C., Lorkovic Z.J., Sasaki T., Naumann U., Matzke A.J., Matzke M. Use of forward genetic screens to identify genes required for RNA-directed DNA methylation in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 2012;77:195–204. doi: 10.1101/sqb.2012.77.015099. [DOI] [PubMed] [Google Scholar]
- 40.Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23(24):2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19(5):1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Ma Z.Y., Zeng L., Tanaka K., Zhang C.J., Ma J., Bai G., Wang P., Zhang S.W., Liu Z.W., Cai T., Tang K., Liu R., Shi X., He X.J., Zhu J.K. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA. 2013;110(20):8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law J.A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7(7):e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498(7454):385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J., Bai G., Zhang C., Chen W., Zhou J., Zhang S., Chen Q., Deng X., He X.J., Zhu J.K. An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res. 2011;21(12):1691–1700. doi: 10.1038/cr.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He X.J., Hsu Y.F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.S., Jin H., Zhu J.K. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137(3):498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10(6):649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowley M.J., Avrutsky M.I., Sifuentes C.J., Pereira L., Wierzbicki A.T. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7(6):e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He X.J., Hsu Y.F., Zhu S., Liu H.L., Pontes O., Zhu J., Cui X., Wang C.S., Zhu J.K. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23(23):2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanno T., Bucher E., Daxinger L., Huettel B., Kreil D.P., Breinig F., Lind M., Schmitt M.J., Simon S.A., Gurazada S.G., Meyers B.C., Lorkovic Z.J., Matzke A.J., Matzke M. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 2010;11(1):65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanno T., Bucher E., Daxinger L., Huettel B., Böhmdorfer G., Gregor W., Kreil D.P., Matzke M., Matzke A.J. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 2008;40(5):670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 52.Gao Z., Liu H.L., Daxinger L., Pontes O., He X., Qian W., Lin H., Xie M., Lorkovic Z.J., Zhang S., Miki D., Zhan X., Pontier D., Lagrange T., Jin H., Matzke A.J., Matzke M., Pikaard C.S., Zhu J.K. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465(7294):106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law J.A., Ausin I., Johnson L.M., Vashisht A.A., Zhu J.K., Wohlschlegel J.A., Jacobsen S.E. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20(10):951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson L.M., Law J.A., Khattar A., Henderson I.R., Jacobsen S.E. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4(11):e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuhlmann M., Mette M.F. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol. Biol. 2012;79(6):623–633. doi: 10.1007/s11103-012-9934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson L.M., Du J., Hale C.J., Bischof S., Feng S., Chodavarapu R.K., Zhong X., Marson G., Pellegrini M., Segal D.J., Patel D.J., Jacobsen S.E. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507(7490):124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z.W., Shao C.R., Zhang C.J., Zhou J.X., Zhang S.W., Li L., Chen S., Huang H.W., Cai T., He X.J. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10(1):e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wassenegger M., Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11(3):142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Zong J., Yao X., Yin J., Zhang D., Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447(1):29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16(10):1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126(1):79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 62.Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126(1):93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 63.Haag J.R., Ream T.S., Marasco M., Nicora C.D., Norbeck A.D., Pasa-Tolic L., Pikaard C.S. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell. 2012;48(5):811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aalto A.P., Poranen M.M., Grimes J.M., Stuart D.I., Bamford D.H. In vitro activities of the multifunctional RNA silencing polymerase QDE-1 of Neurospora crassa. J. Biol. Chem. 2010;285(38):29367–29374. doi: 10.1074/jbc.M110.139121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13(7):350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 66.Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22(2):321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye R., Wang W., Iki T., Liu C., Wu Y., Ishikawa M., Zhou X., Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell. 2012;46(6):859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 68.Ausin I, Greenberg MV, Simanshu DK, Hale CJ, Vashisht AA, Simon SA, Lee TF, Feng S, Espanola SD, Meyers BC, Wohlschlegel JA, Patel DJ, Jacobsen SE. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Nat. Acad. Sci. U.S.A. . 2012;109(22):8374–81. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ausin I., Mockler T.C., Chory J., Jacobsen S.E. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16(12):1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie M., Ren G., Zhang C., Yu B. The DNA- and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J. 2012;72(3):491–500. doi: 10.1111/j.1365-313X.2012.05092.x. [DOI] [PubMed] [Google Scholar]
- 71.Xie M., Ren G., Costa-Nunes P., Pontes O., Yu B. A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res. 2012;40(10):4422–4431. doi: 10.1093/nar/gks034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng Z., Xing Y., He X.J., Li W., Hu Y., Yadav S.K., Oh J., Zhu J.K. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J. 2010;62(1):92–99. doi: 10.1111/j.1365-313X.2010.04130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang C.J., Ning Y.Q., Zhang S.W., Chen Q., Shao C.R., Guo Y.W., Zhou J.X., Li L., Chen S., He X.J. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet. 2012;8(5):e1002693. doi: 10.1371/journal.pgen.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mourrain P., Béclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Jouette D., Lacombe A.M., Nikic S., Picault N., Rémoué K., Sanial M., Vo T.A., Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101(5):533–542. doi: 10.1016/S0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 75.Böhmdorfer G., Rowley M.J., Kuciński J., Zhu Y., Amies I., Wierzbicki A.T. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014;79(2):181–191. doi: 10.1111/tpj.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y., Rowley M.J., Böhmdorfer G., Wierzbicki A.T. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013;49(2):298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao X., Jacobsen S.E. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12(13):1138–1144. doi: 10.1016/S0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 78.Bernatavichute Y.V., Zhang X., Cokus S., Pellegrini M., Jacobsen S.E. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3(9):e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enke R.A., Dong Z., Bender J. Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana. PLoS Genet. 2011;7(10):e1002350. doi: 10.1371/journal.pgen.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 81.Ebbs M.L., Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18(5):1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012;158(1):119–129. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.To T.K., Kim J.M., Matsui A., Kurihara Y., Morosawa T., Ishida J., Tanaka M., Endo T., Kakutani T., Toyoda T., Kimura H., Yokoyama S., Shinozaki K., Seki M. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7(4):e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deleris A., Greenberg M.V., Ausin I., Law R.W., Moissiard G., Schubert D., Jacobsen S.E. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 2010;11(12):950–955. doi: 10.1038/embor.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Searle I.R., Pontes O., Melnyk C.W., Smith L.M., Baulcombe D.C. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010;24(10):986–991. doi: 10.1101/gad.579910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greenberg M.V., Deleris A., Hale C.J., Liu A., Feng S., Jacobsen S.E. Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 2013;9(11):e1003946. doi: 10.1371/journal.pgen.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447(7145):735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 88.Moissiard G., Cokus S.J., Cary J., Feng S., Billi A.C., Stroud H., Husmann D., Zhan Y., Lajoie B.R., McCord R.P., Hale C.J., Feng W., Michaels S.D., Frand A.R., Pellegrini M., Dekker J., Kim J.K., Jacobsen S.E. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336(6087):1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moissiard G., Bischof S., Husmann D., Pastor W.A., Hale C.J., Yen L., Stroud H., Papikian A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E. Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc. Natl. Acad. Sci. USA. 2014;111(20):7474–7479. doi: 10.1073/pnas.1406611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013;162(1):116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013;45(9):1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 92.Pontier D., Picart C., Roudier F., Garcia D., Lahmy S., Azevedo J., Alart E., Laudié M., Karlowski W.M., Cooke R., Colot V., Voinnet O., Lagrange T. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell. 2012;48(1):121–132. doi: 10.1016/j.molcel.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 93.Garcia D., Garcia S., Pontier D., Marchais A., Renou J.P., Lagrange T., Voinnet O. Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol. Cell. 2012;48(1):109–120. doi: 10.1016/j.molcel.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 94.Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. DNA methylation mediated by a microRNA pathway. Mol. Cell. 2010;38(3):465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 95.Wu L., Mao L., Qi Y. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 2012;160(2):990–999. doi: 10.1104/pp.112.200279. [DOI] [PMC free article] [PubMed] [Google Scholar]