Fig. 3.
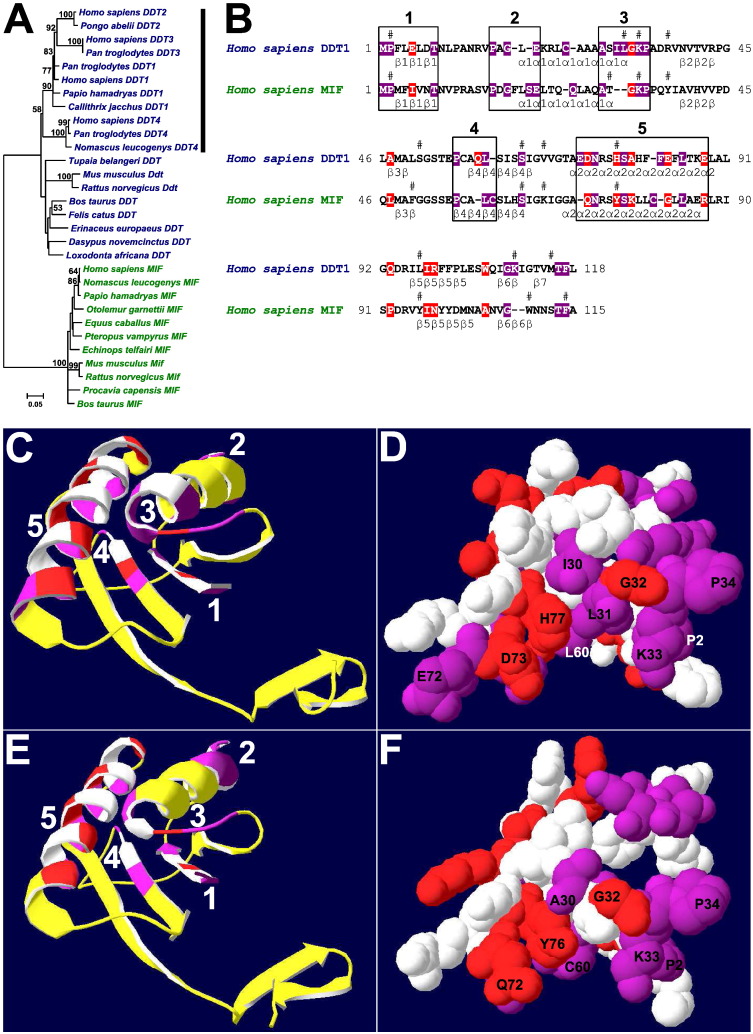
Analysis of eutherian D-dopachrome tautomerase and macrophage migration inhibitory factor genes. (A) Minimum evolution tree of eutherian D-dopachrome tautomerase and macrophage migration inhibitory factor genes. The tree was calculated using maximum composite likelihood method. The estimates > 50% were shown, after 1000 bootstrap replicates. The vertical bar labelled primate-specific differential gene expansions DDT1-4. (B–F) Protein molecular evolution analysis. (B) Reference human DDT1 and MIF protein amino acid sequences. The invariant amino acid sites were shown using white letters on violet backgrounds. The forward amino acid sites were shown using white letters on red backgrounds. The amino acid clusters 1–5 with overrepresented invariant or/and forward amino acid sites were labelled using rectangles. The secondary structure elements were designated according to Sugimoto et al. (1999) for DDT1 crystal structure 1DPT, and according to Sun et al. (1996) for MIF crystal structure 1MIF. The amino acid residues implicated in putative DDT1 and MIF active sites (Sugimoto et al., 1999) were labelled by #s. (C–D) Analysis of human DDT1 crystal structure 1DPT. (C) Ribbon representation of human DDT1 crystal structure 1DPT. (D) van-der-Waals representations of amino acid cluster 1-5 amino acids in view identical to that in C. (E–F) Analysis of human MIF crystal structure 1MIF. (E) Ribbon representation of human MIF crystal structure 1MIF. (F) van-der-Waals representations of amino acid cluster 1-5 amino acids in view identical to that in E. (C–F) The amino acid cluster 1-5 invariant amino acid sites were labelled violet, forward amino acid sites were labelled red and compensatory amino acid sites were labelled white.