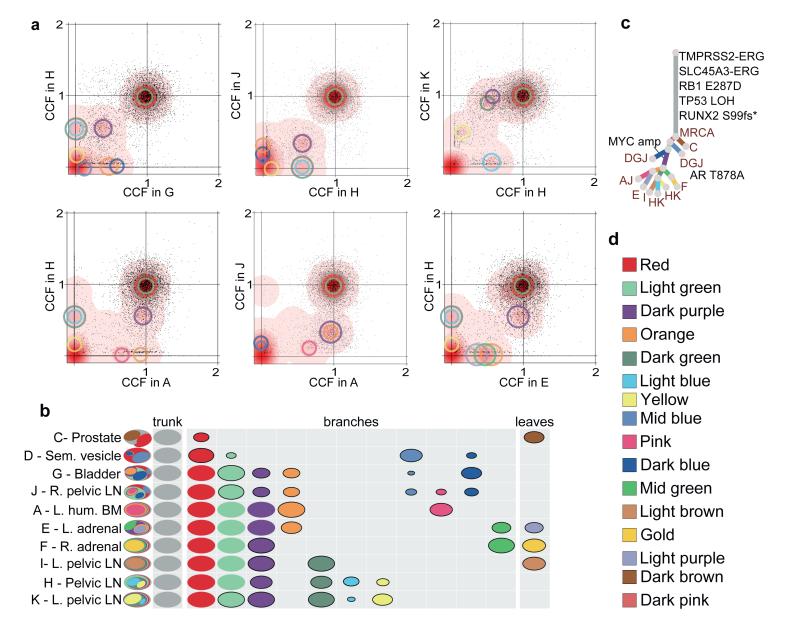
Figure 1. n-D Dirichlet process clustering reveals widespread polyclonal seeding in A22.
(a) For pairs of metastases, cancer cell fractions (CCF), i.e. the fraction of cells within a sample containing a mutation, are plotted for all the substitutions detected in the WGS data. Red density areas off the axes and with CCF >0 and <1 reveal the existence of mutation clusters present at subclonal levels in more than one metastatic site. Mutation clusters for each sample are indicated with circles coloured according to the subclone they correspond to (Supplementary Table 3). The centre of each circle is positioned at the CCF values of the subclone in the two samples. The clusters at (1,1) correspond to the mutations present in all the cells in both sites (CCF=1) while those on axes refer to sample-specific subclones. For example, light blue and dark green clusters absent from sample A are positioned on the y-axis when H is compared to A but are moved to (0.60,0.08) and (0.60,0.88) when H is compared to K. (b) Each subclone detected in A22 is represented as a set of colour-coded ovals across all organ sites (Supplementary Table 3). Each row represents a sample, with ovals in the far left column nested if required by the pigeonhole principle (SI). The area of the ovals is proportional to the CCF of the corresponding subclone. Subclonal mutation clusters are shown with solid borders. Oval plots are divided into three types: trunk (CCF=1 in all samples), leaf (specific to a single sample) and branch (present in >1 sample and either not found in all samples or subclonal in at least one). (c) Phylogenetic tree showing the relationships between subclones in A22. Branch lengths are proportional to the number of substitutions in each cluster. Branches are annotated with samples in which they are present and with oncogenic/putative oncogenic alterations assigned to that subclone (LOH: Loss of Heterozygosity). (d) Subclone colour key.