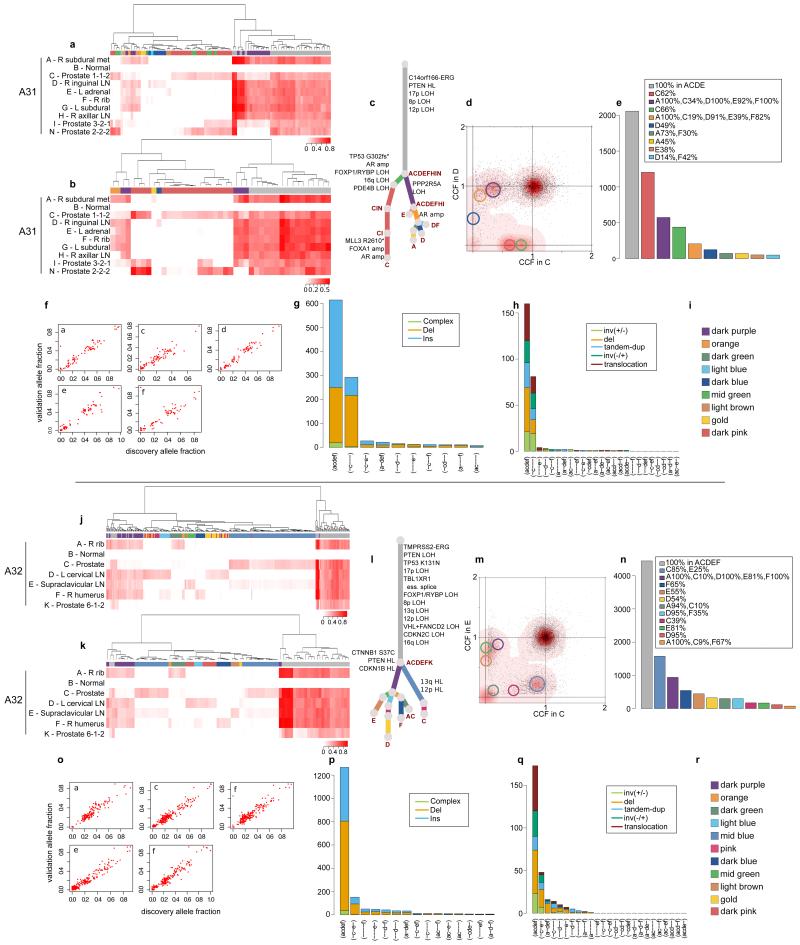
Extended Data Figure 3. Validation of the subclonal hierarchies in A31 and A32.
Validation strategy as described in Extended Data Figure 2. For A31 and A32, hierarchical clustering of the VAF was performed separately for substitutions (a) and (j) and indels (b) and (k). Heatmaps are annotated as described in Extended Data Figure 2. Additional samples for A31 and A32 are incorporated into the phylogenetic trees (c) and (l). Subclones for A31 CD and A32 CE are annotated in the corresponding 2d-DP plots (d) and (m). Numbers of substitutions in WGS data assigned to each subclone are plotted in (e) and (n). VAFs from WGS and validation data, plotted as scatter plots (f) and (o), are very highly correlated. Number of indels (g) and (p) and rearrangements (h) and (q) present in different subsets of samples are plotted as bar charts. Subclone Colour keys for A31 and A32 (i and r) respectively.