Fig 2.
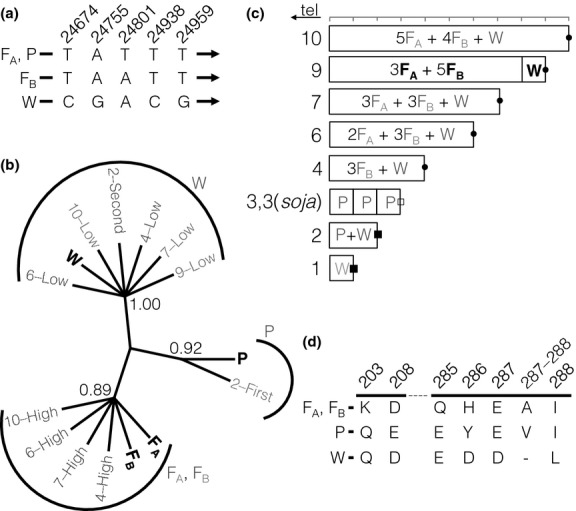
Sequence of the Rhg1 repeat units. (a) Five examples of sequence variants used for reconstruction of repeat units in an intergenic region between Glyma18g02610 and Glyma18g02620 are displayed. Three patterns in these five nucleotides differentiate three of four separate repeat units (FA, a repeat unit found in the PI 88788 genotype; P, a repeat unit found in the Peking genotype; FB, another repeat unit found in the PI 88788 genotype; and W, a single-copy version of the sequence found in the susceptible Williams 82 genotype). Positions are given relative to the first nucleotide (1 632 225 bp on chromosome 18) of the 31.2-kb repeat in the Williams 82 genome assembly. (b) Classification of repeat units using maximum parsimony analysis of sequences. Reconstructed individual Glyma18g02590 genes from the repeats are labelled according to copy number and relative abundance in the accession (e.g. ‘4-low’ means the less abundant sequence present in a four-copy genotype) or by position relative to the telomere if equally abundant (e.g. 2-first). Bootstrap support values are given above key nodes. (c) Interpretation of the Rhg1 repeat structure. Bold black labels represent sequences with position known from large insert cloning; grey labels are inferred from short-read shotgun sequence data classified by the parsimony analysis in ‘b’. Rhg1 copy number in the Glycine max accession genome is denoted on the left. Three different fusion sequences at the centromere-proximal end are marked by open squares, filled squares and filled circles. tel: telomere. (d) Amino acid variation in the predicted α-SNAP protein, Glyma18g02590. Amino acid positions are from the Williams 82 reference. Bold lines represent exons 6 and 9, respectively.
