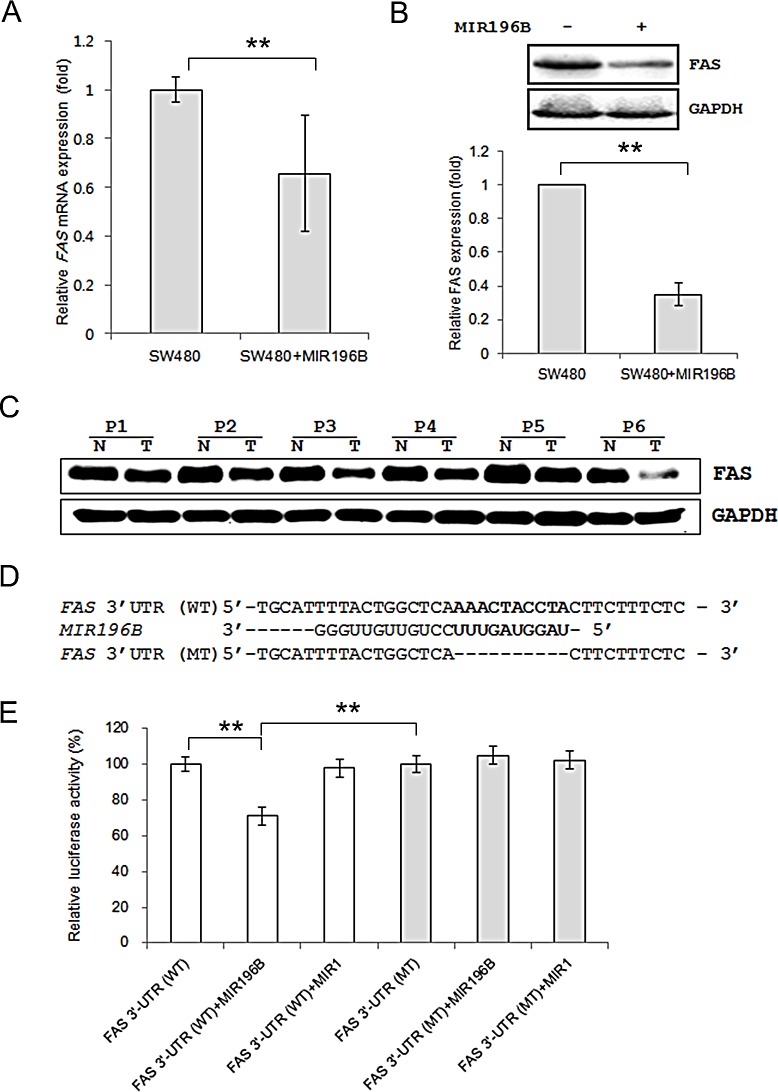
Figure 2. FAS is a direct target gene of MIR196B.
(A) qRT-PCR analysis of FAS expression in SW480 cells. SW480 cells were mock-transfected or transfected with pre-MIR196B. Cells were harvested 48 h after transfection. Total RNA was extracted and used for qRT-PCR. Values are presented as the fold-change in MIR196B-overexpressing cells relative to non-transfected cells. The experiment was performed in duplicate and repeated five times. (B) FAS protein levels in MIR196B-overexpressing cells and non-transfected cells. Protein was extracted 72 h after transfection for western blot analysis. (C) FAS expression in 6 pairs of colon cancer tissue samples and adjacent normal colon tissue samples. All samples are from patients with colon cancer. (D) Sequence alignment of the wild-type (WT) and mutated (MT) MIR196B target site in the 3′-UTR of FAS. A human FAS 3′ UTR containing the wild-type and mutant MIR196B binding sequence was cloned downstream of the luciferase reporter gene. (E) A luciferase reporter plasmid containing the WT or MT FAS 3′ UTR was co-transfected into SW480 cells with pre-MIR1 as a negative control or pre-MIR196B. Luciferase activity was determined using the dual luciferase assay. Results are shown as the relative firefly luciferase activity normalized to Renilla luciferase activity. Data assessed from three independent experiments and the P values were calculated by t-test (* P < 0.05; ** P < 0.01).