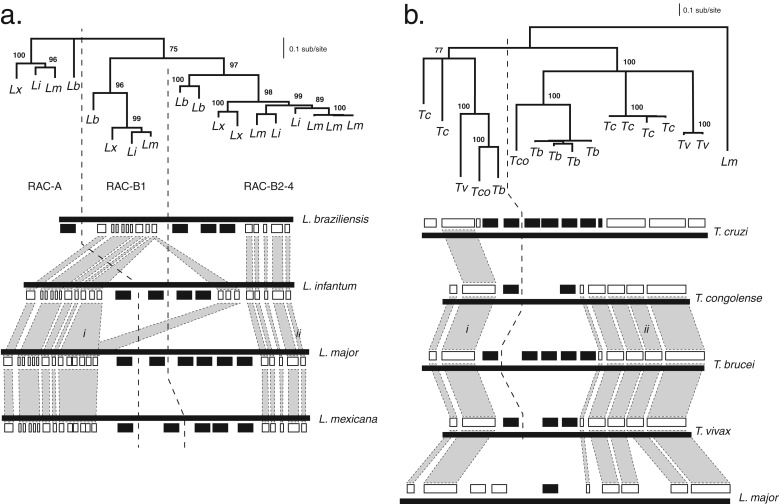
Fig. 4.
Structural differentiation of paralogues in tandem gene arrays. a. Receptor-type adenylate cyclase (rac) genes in four Leishmania species. The rac array (i.e. LmjF.17·0200) is located at the extreme left-end of chromosome 17 in L. major; its conserved position is defined downstream by an EF1α gene array (i) and upstream by a metalloprotease (ii). The structure of the array in four species is depicted, rac genes are shown in black, and other loci are shown in white. Vertical grey shading represents homology between flanking loci, to demonstrate positional conservation. The dashed black line separates array positions that correspond with distinct clades in the phylogeny. A maximum likelihood phylogeny estimated from amino acid sequences using a LG+Γ model is shown, with non-parametric bootstraps applied to nodes where support is >75. Terminal nodes are labelled with species name initials. The tree is midpoint rooted. b. Cation transporter genes in four Trypanosoma species. The transporter array (i.e. Tb927.11.9000) is located on chromosome 11 in T. brucei; its conserved position is defined downstream by a palmitoyl acyltransferase 4 gene (i) and upstream by an EF1γ2 gene (ii). The phylogeny and genome comparison are as depicted in a. except that the tree is rooted with a single-copy orthology from L. major.