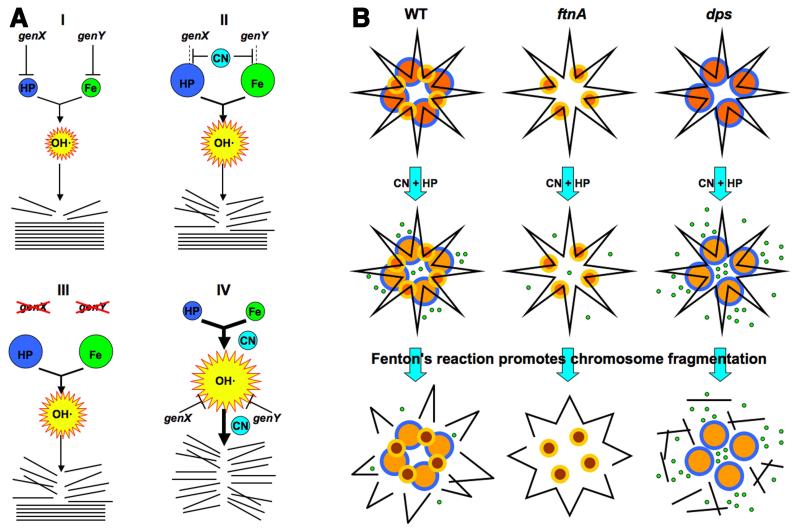
Fig. 7. The interpretation scheme and the chromosome fragmentation scheme.
A. Interpretation of genetic results. The intracellular HP, iron and OH· pools are shown by spheres whose size reflect the relative size of the pools. Stack of straight lines at the bottom, chromosomal DNA; broken lines, broken DNA. (I) Our initial HP-only treatment scenario, in which both HP and iron pools are actively limited by dedicated cellular functions, genX and genY. (II) Our initial expectation of a typical CN-potentiation route (inactivation of the HP- or iron-limiting functions). (III) The expected phenotypes of the corresponding mutants (in functions that are CN-targets) in HP-only treatment. (IV) An alternative scenario suggested by the most frequently observed phenotypes of mutants (no HP-only sensitivity, increased CN+HP sensitivity), according to which CN stimulates Fenton’s reaction near DNA. B. The proposed role of ferritin-like proteins in CN potentiation of HP poisoning via catastrophic chromosome fragmentation, based on phenotypes of the corresponding mutants. Top row: yellow/orange smaller circles, Dps dodecamers; blue/orange bigger circles, ferritin 24-mers; black line in the shape of a star, chromosomal (duplex) DNA. Middle row: small green circles, released ferrous iron (note a lighter color of the ferritin circles). In the absence of ferritin, some ferrous iron is still coming from other (unknown) sources. Bottom row: broken black lines, chromosome fragments. The released iron is taken into Dps spheres (note their darker color). For clarity, DNA fragments are shown dissociated from ferritin and Dps.