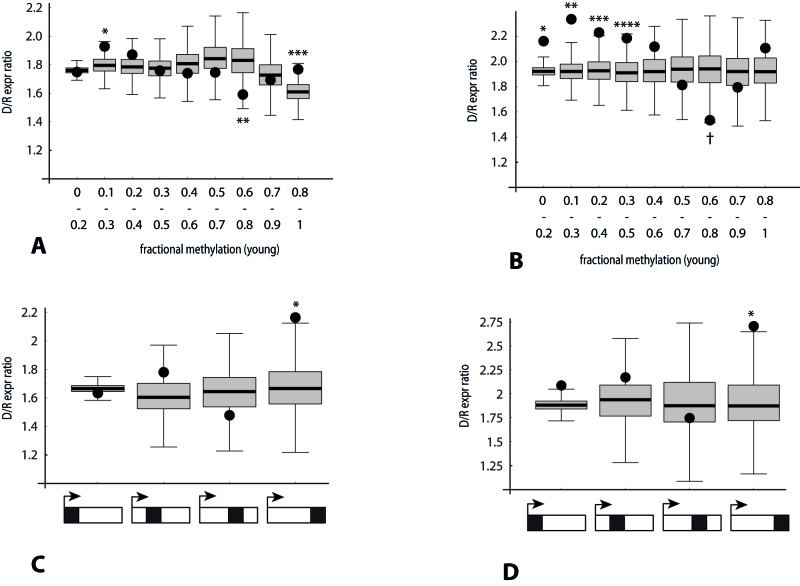
Figure 8. Association of the direction of gene expression change with bisulfite peak density and methylation levels in RRBS data (analysis 2).
Y-axis: Ratio (number of genes with decreased expression in poor responders: number of genes with increased expression). Black dots: median observed ratio in each category. Box and whisker: distribution of ratios expected by chance in each category, obtained by repeated (10,000-fold) random sampling of two reference gene set pools (lower and higher bsDens) in order to approximate the observed bsDens for that category. See legend to Figure 5 for derivation of reference gene set (n = 15,203). p-values: fraction of sampling-derived ratios with greater deviation from the median than the observed ratio for that category. (A) Ten overlapping categories defined by average methylation level of RRBS loci within the gene (binned X-axis value ranges 0 to 0.2, 0.1 to 0.3, 0.2 to 0.4, etc.).*p < 0.02; **p < 0.019; ***p < 0.025. (B) Same as (A), except analysis restricted to genes with RRBS peak density > 0.5. *p < 0.0001; **p < 0.0001; ***p < 0.003; ****p < 0.012; †p < 0.0016. (C) Four categories depicted below X-axis indicate RRBS read loci localized within (left to right): quartile of gene length proximal to 5ʹ-end, second quartile, third quartile, or quartile of gene length proximal to 3ʹ-end. p-value for 3ʹ-end quartile < 4.5 × 10−3. (D) Same as (C), except analysis restricted to genes with RRBS peak density > 0.5. p-value for 3ʹ-end quartile < 0.011.