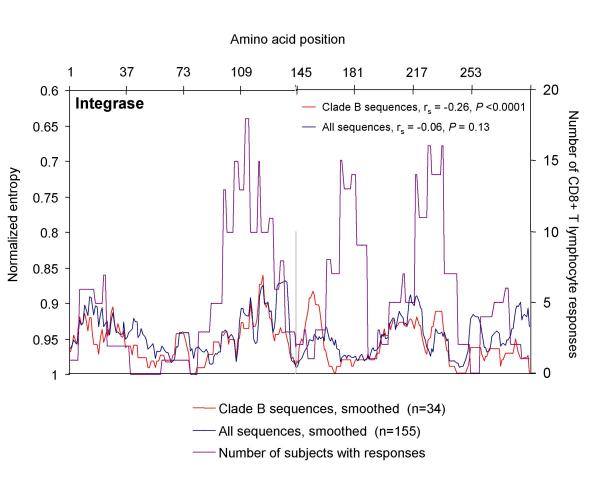
Figure 5.
Correlation of amino acid sequence variability with frequency of CD8 T cell responses targeting Integrase. For Integrase, amino acid sequences were obtained from at the HIV-1 Molecular Immunology Database (27), and aligned relative to the HIV-1 clade B consensus sequence. Entropy scores for each amino acid residue were calculated based on this alignment, smoothed over nine amino acids, and plotted for all sequences (n = 155, blue line, left axis) and clade B sequences only (n = 34, red line, left axis). Entropy scores of 1 correspond to 100% conserved residues, while lower scores (plotted here on an inverse scale) correspond to increasing sequence variability. The number of responses in the 56 study subjects against peptides containing each amino acid was also plotted (purple line, right axis) to correlate regions with high sequence variability with regions targeted by CD8 T cells. Spearman's rank-order correlation coefficient was calculated to correlate CD8 T cell responses against sequence variability for each protein.