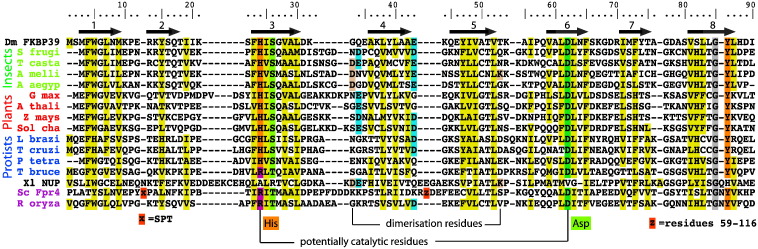
Fig. 2.
Structural alignment of NPL sequences. The structures of Xenopus nucleoplasmin (1KJ5, labelled “Xl NUP”) and FKBP39 were aligned and the other sequences were added based on sequence only. The two other proteins used in our experiments are yeast Fpr4 and A. thaliana HD2 (“A thali”). The residue numbering and the position of β-strands of FKBP39 are indicated above its sequence. The positions of the conserved and potentially catalytic histidine and aspartate are indicated, as well as the two dimer-promoting residues of nucleoplasmin. The yeast protein Fpr4, also an NPL-FKBP, contains a large insertion (residues 59–116, indicated by “z”) that has been omitted from the alignment. The other proteins are from the fungus Rhizopus oryzae; the insects Spodoptera frugiperda, Tribolium castaneum, A. mellifera and Aedes aegypti; the plants Glycine max, A. thaliana, Zea mays and Solanum chacoense; and the protozoa Leishmania braziliensis, Trypanosoma cruzi, Paramecium tetraurelia and Trypanosoma brucei.