Abstract
Background
Starch is biosynthesised by a complex of enzymes including various starch synthases and starch branching and debranching enzymes, amongst others. The role of all these enzymes has been investigated using gene silencing or genetic knockouts, but there are few examples of overexpression due to the problems of either cloning large genomic fragments or the toxicity of functional cDNAs to bacteria during cloning. The aim of this study was to investigate the function of potato STARCH BRANCHING ENZYME II (SBEII) using overexpression in potato tubers.
Results
A hybrid SBEII intragene consisting of potato cDNA containing a fragment of potato genomic DNA that included a single intron was used in order to prevent bacterial translation during cloning. A population of 20 transgenic potato plants exhibiting SBEII overexpression was generated. Compared with wild-type, starch from these tubers possessed an increased degree of amylopectin branching, with more short chains of degree of polymerisation (DP) 6–12 and particularly of DP6. Transgenic lines expressing a GRANULE-BOUND STARCH SYNTHASE (GBSS) RNAi construct were also generated for comparison and exhibited post-transcriptional gene silencing of GBSS and reduced amylose content in the starch. Both transgenic modifications did not affect granule morphology but reduced starch peak viscosity. In starch from SBEII-overexpressing lines, the increased ratio of short to long amylopectin branches facilitated gelatinisation, which occurred at a reduced temperature (by up to 3°C) or lower urea concentration. In contrast, silencing of GBSS increased the gelatinisation temperature by 4°C, and starch required a higher urea concentration for gelatinisation. In lines with a range of SBEII overexpression, the magnitude of the increase in SBEII activity, reduction in onset of gelatinisation temperature and increase in starch swollen pellet volume were highly correlated, consistent with reports that starch swelling is greatly dependent upon the amylopectin branching pattern.
Conclusion
This work reports the first time that overexpression of SBEII has been achieved in a non-cereal plant. The data show that overexpression of SBEII using a simple single-intron hybrid intragene is an effective way to modify potato starch physicochemical properties, and indicate that an increased ratio of short to long amylopectin branches produces commercially beneficial changes in starch properties such as reduced gelatinisation temperature, reduced viscosity and increased swelling volume.
Electronic supplementary material
The online version of this article (doi:10.1186/s12896-015-0143-y) contains supplementary material, which is available to authorized users.
Keywords: Amylopectin branching, GBSS silencing, SBEII overexpression, Starch gelatinisation, Starch properties, Solanum tuberosum
Background
Starch serves as the most important carbohydrate storage reserve for plants, and provides the foundation for human and animal nutrition as well as a raw material for numerous food and industrial applications [1]. Starch consists of two molecules, long essentially linear chains of α-(1 → 4)-linked glucopyranosyl residues (amylose), and a much larger, highly branched molecule (amylopectin) consisting of thousands of short α-(1 → 4)-linked glucan chains of degree of polymerisation (DP) 6 to >100 that are attached by α-(1 → 6)-linked branch points [2].
Amylose and amylopectin are synthesised in the plastids, where they assemble into a semicrystalline granule. Amylopectin is the major component, making up 70–80% of starch in most species. Amylose is synthesised by granule-bound starch synthase (GBSS), whereas a large complex of enzymes is required to synthesise amylopectin. This complex consists of four soluble starch synthases (SSI, SSII, SSIII, SSIV) and two types of starch branching enzyme (SBEI, SBEII), with various debranching enzymes, kinases and other enzymes also involved [2-4]. The availability of various mutants lacking particular enzyme activities has shown how the properties of starch from different species are altered by changes to composition or structure. For example, starch from the waxy mutant of maize (Zea mays L.) (deficient in GBSS and virtually amylose-free) had a lower gelatinisation temperature, whereas that from wheat (Triticum aestivum L.) had an unaltered gelatinisation temperature but altered pasting properties [5]. Starch from potato (Solanum tuberosum L.) where GBSS had been down-regulated produced gels with improved stability and clarity [6].
Differences in amylopectin branching affect granule crystallinity, which together with differences between species in granule size and shape result in altered thermal, pasting and biophysical properties [7,8]. Amylopectin branching is carried out by SBEI and SBEII, enzymes that create branch points by cleaving α-(1 → 4)-linkages and reattaching the glucan chain via an α-(1 → 6)-linkage. Alterations in SBE activity affect the number and size distribution of amylopectin branches. In cereals, there are three isoforms of SBE: SBEIa, SBEIIa and SBEIIb, and in the amylose extender mutant of maize that lacks SBEIIb, amylopectin had fewer branches and a higher proportion of longer chains [7] that are essentially amylose. Similarly in rice (Oryza sativa L.), down-regulation of SBEIIb decreased the proportion of short branches and increased the proportion of intermediate and long branches [9]. In barley (Hordeum vulgare L.), only when both SBEIIa and SBEIIb were down-regulated was a significant reduction in the frequency of amylopectin branches observed [10]. Indeed, down-regulation of all three isoforms of SBE in barley produced starch that was entirely amylose [11]. In vitro experiments with rice enzymes found that SBEIIb transferred chains of DP6 and DP7, SBEIIa transferred chains of DP6–15, and SBEIa produced a range of short and intermediate chains of DP ≤ 40 [12].
In potato only two isoforms of SBE are involved, SBEI (also known as SBE B) and SBEII (also known as SBE A) [13,14]. SBEI is the major isoform, but its down-regulation had little effect on starch structure [15,16]. Knockdown of the minor SBEII isoform, however, increased the average chain length of amylopectin, which, as in the maize amylose extender mutant [17], resulted in an increase in apparent amylose content due to the increased proportion of longer linear amylopectin chains [14]. Knockdown of both SBEI and SBEII produced potato starch that was >70% apparent amylose and had a very high phosphorus content [18].
Overexpression of SBE activity to modify starch structure is a much more difficult proposition than knockdown, due to the enzyme’s deleterious effects on bacteria during cloning of the constructs [14]. Overexpression of bacterial glycogen synthase or glycogen branching enzyme produced starch with more branches and more short chains [19-21], but only one study has successfully overexpressed a plant SBE enzyme. This involved the cloning of an 18 kb genomic fragment containing the native promoter and coding region, including 21 introns, of the rice SBEIIb gene, and expressing it in a rice SBEIIb-deficient background [22]. We aimed to examine the role of potato SBEII in starch branching and resultant effects on potato starch properties. Potato is now the third most important food crop in the world and the most important non-cereal [23], whose starch also has many uses in the food and manufacturing industries [1] due to its advantages over cereal starches (see Discussion). To do this we employed a simple hybrid cDNA/gDNA intragene construct containing a single intron to prevent bacterial translation, expressed under the control of the strong tuber-specific potato GBSS promoter. An additional way to increase the proportion of branched amylopectin in the granule is to down-regulate GBSS and reduce amylose content, so knockdown GBSS lines were also generated for comparison.
Results
Characterisation of transgenic potato plants
Initial experiments confirmed an earlier report that a complete cDNA of potato SBEII was unable to be propagated in Escherichia coli [14]. Despite the use of several different bacterial strains, very few colonies were produced and all were subsequently discovered to contain frame-shift mutations (data not shown). Therefore, a complete but hybrid SBEII intragene containing a single intron to prevent bacterial translation was assembled from cDNA and genomic DNA fragments (See Methods and Additional file 1: Figure S1). A population of 28 plants transformed with the SBEII-intron intragene under the control of the GBSS promoter was generated (Additional file 1: Figure S2). Of these, 20 plantlets showing good root development were grown to maturity, and tubers exhibited a range of SBEII mRNA abundances (Figure 1A). SBEII mRNA accumulation in these lines ranged from less than wild-type controls (line 1047–3), moderate overexpression (lines 1047–1, 2, 4, 5, 6, 7, 8, 9, 11, 14, 18, 20, 22, 24) to high overexpression (lines 1047–12, 15, 17, 23). Line 1047–19 exhibited approximately wild-type mRNA abundance of SBEII but with an additional anomalous larger SBEII transcript presumably derived from an aberrant insertion event. To confirm that mRNA accumulation of the structurally unrelated starch synthase enzymes SSII and SSIII was not affected by manipulation of SBEII expression, the same gel blot was stripped and re-hybridised. Sequential re-hybridisation with labelled probes for SSII and SSIII showed that the mRNA abundances of these two genes were essentially indistinguishable from wild-type and empty vector controls (Additional file 1: Figure S3).
Figure 1.
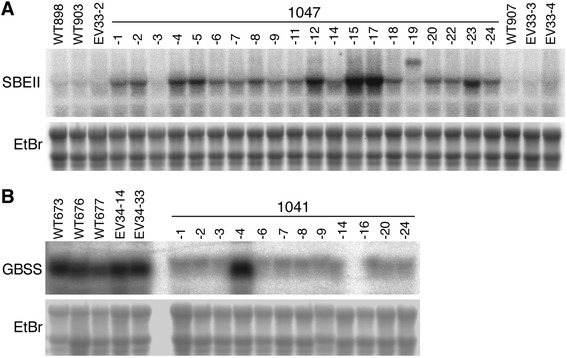
RNA gel blots showing alterations to SBEII and GBSS mRNA abundance in transgenic plants. (A) SBEII mRNA abundance in tubers from plants transformed with a SBEII overexpression construct (1047 lines). (B) GBSS mRNA abundance in tubers from plants transformed with a GBSS RNAi construct (1041 lines). Wild-type (WT) and empty vector transformants (EV) are shown for comparison. Ethidium bromide (EtBr) staining of RNA (with contrast reversed) was used to confirm the equivalency of loading.
Of a population of 24 plants putatively transformed with a GBSS inverted repeat transgene, only 12 were positive for the presence of both the transgene and the selectable marker gene (Additional file 1: Figure S2). Of these 12 lines, 11 showed a strong reduction in GBSS mRNA accumulation (Figure 1B). One line, 1041–4, did not show any apparent posttranscriptional gene silencing, and this line was retained as an additional control for some tests since it was transgenic but GBSS mRNA accumulation was not reduced relative to wild-type.
To assess the effect of overexpression of a SBEII intragene on SBE enzyme activity, SBEII was separated from the major and predominant SBEI by separating proteins using native polyacrylamide gel electrophoresis (PAGE), followed by an in-gel activity reaction and staining for SBE reaction products (Figure 2A). The two isoforms of SBE were identified based on the migration of rice and wheat enzymes [17,22,24] and the relative activities of potato SBEI and SBEII [14]. SBEII activity approximately reflected the abundance of SBEII mRNA, being similar to wild-type in line 1047–3, and with a range of activities greater than wild-type in a selection of other lines (WT < 1047–5 < 1047–23 = 1047–12 ≤ 1047–17 ≤ 1047–15). The greatest enhancement was in lines 1047–15 and 17, which possessed SBEII activity almost 3-fold that of wild-type (Figure 2B).
Figure 2.
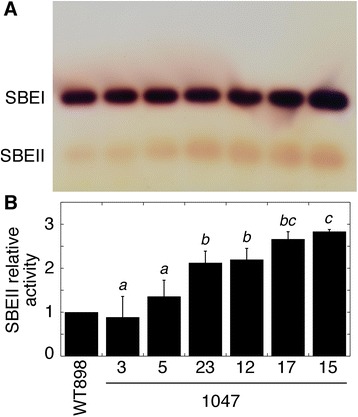
SBE activity in tubers from a series of lines transformed with a SBEII overexpression construct. (A) Native PAGE activity gel with SBE activity visualized by iodine staining. (B) Mean relative SBEII activity ± SD as determined from the density of the spots. Data are from technical replicate protein preparations from the same pooled tissue as Figure 1 run on different gels on different days (n = 3). Means not sharing a common letter are significantly different between groups at P = 0.05 as determined by LSD after a one-way ANOVA test.
Effects of altered GBSS and SBEII expression on the structure of starch
The effectiveness of silencing of GBSS was determined by measuring the percentage of amylose present in the starch from these plants (Table 1). Wild-type and empty vector control starches were approximately 19–26% amylose, but this was reduced to 3.1–5.6% in lines 1041–1, 3 and 16. In line 1041–4, which showed no reduction in GBSS mRNA abundance relative to controls (Figure 1B), amylose comprised 21.3% of the total starch, a content within the range found in controls. The percentage of amylose in the 1047 lines overexpressing SBEII ranged from 17.0 to 23.4%, and was similar to controls.
Table 1.
Amylose content in starch from potato tubers of a range of lines exhibiting silencing of GBSS (1041 lines) or overexpressing SBEII (1047 lines)
| Line | % Amylose |
|---|---|
| WT676 | 26.3 ± 0.3 |
| WT898 | 21.4 ± 0.3 |
| WT903 | 21.5 ± 0.9 |
| EV33-3 | 20.6 ± 0.2 |
| EV33-5 | 18.8 ± 0.8 |
| EV34-33 | 22.2 ± 0.2 |
| 1041-1 | 4.9 ± 0.3 |
| 1041-3 | 3.1 ± 1.0 |
| 1041-4 | 21.3 ± 0.2 |
| 1041-16 | 5.6 ± 0.3 |
| 1047-3 | 19.2 ± 0.3 |
| 1047-5 | 17.0 ± 0.3 |
| 1047-12 | 17.9 ± 0.6 |
| 1047-15 | 23.4 ± 0.8 |
| 1047-17 | 19.4 ± 0.3 |
| 1047-23 | 22.2 ± 0.8 |
Starch was also prepared and assayed from wild-type (WT) and empty vector (EV) control plants grown alongside the transgenic lines. Data are means ± SD (n = 4 technical replicates).
Size exclusion chromatography was used to separate amylose from amylopectin (Figure 3). Elution profiles consisted of two main regions, a sharp peak eluting at 9–11 min representing high molecular weight amylopectin, and a broader more disperse lower molecular weight peak eluting at 12–17 min representing amylose [25-27]. The small peaks eluting at 18–19 min were probably due to free sugars. The relative areas of the amylopectin and amylose peaks showed that suppression of GBSS in line 1041–3 substantially reduced the content of amylose and that overexpression of SBEII in line 1047–17 slightly reduced the content of amylose, consistent with the biochemical assays (Table 1). The amylopectin peaks for both transgenic lines eluted at slightly higher molecular weight than wild-type.
Figure 3.
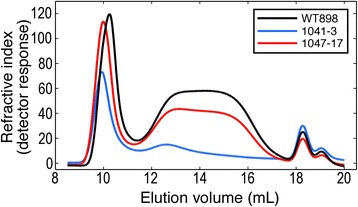
Size separation of tuber starch polyglucans. Starch was solubilised in NaOH and gelatinised by heat before size exclusion chromatography on a Waters Ultrahydrogel Linear column. Curves are the means of measurements on three independent biological replicates per line. Carbohydrate in the eluate was measured by refractive index.
A modified fluorophore-assisted carbohydrate electrophoresis (FACE) method was used to examine the effects of altered SBEII and GBSS expression on the chain length distribution of debranched polyglucans (Figure 4). Polyglucans from wild-type showed a moderate number of side chains of DP6, few side-chains of DP7-9, and a predominant peak of side chains spanning the DP ~ 10-24 range (Figure 4A, C). The number of side chains of DP ≥ 37 was small. Overexpression of SBEII in various lines had little effect on the chain length distribution of moderately sized chains of DP13-18, slightly reduced the abundance of larger chains of DP ≥ 20 and increased the number of short chains of DP ≤ 12 (Figure 4B). The greatest effect was on the number of chains of DP6, with the increase in lines 1047–5, 12, 15, 17 and 23 ranging from 19 to 56% greater than wild-type (Figure 4A). However, there was no obvious correlation between the increase in DP6 chain abundance and relative SBEII activity. The only line in which an increase in the abundance of DP6 side chains was not observed was line 1047–3 (Figure 4B), which was the line in which SBEII mRNA abundance and activity were not increased (Figures 1 and 2).
Figure 4.
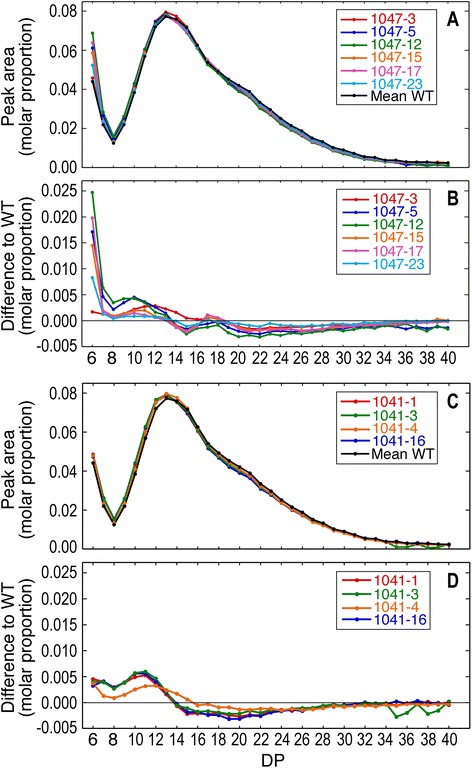
Chain length distribution of amylopectin unit chains. (A) Comparison of size distribution of amylopectin unit chains in starch from tubers with altered SBEII expression. (B) Difference plot showing changes in chain length distribution relative to wild-type. (C) Comparison of size distribution of amylopectin unit chains in starch from tubers with altered GBSS expression. (D) Difference plot showing changes in chain length distribution relative to wild-type.
In transgenic lines with a higher proportion of amylopectin in the granule due to reduced amylose content (lines 1041–1, 3, 16), the chain length distribution of polyglucans was very similar to wild-type (Figure 4C). There was a small increase in the number of side chains of DP6-13, and a small decrease in those of DP15-28 (Figure 4D), changes that were much less pronounced in line 1041–4 that did not exhibit silencing of GBSS (Figure 1). This suggests that the reduced activity of GBSS not only increased the proportion of amylopectin in starch (by reducing the biosynthesis of amylose), but also had some small effects on its structure.
Amylopectin chain length distribution and the phosphorus content of the starch are related, with phosphorylation predominantly occurring on longer amylopectin chains [28]. Many studies of starch phosphorylation underestimate the true abundance of phosphate groups by measuring only C6-linked phosphates, and ignoring the more acid-labile C3 linkage. We have used an improved method that allows determination of phosphate groups with both linkages. The ratio of C6 phosphate content to C3 phosphate content was ~4.5:1 in all lines tested (Table 2). Total phosphate content was ~40 nmol per mg starch in a wild-type line, an empty vector transformant and two lines with knockdown of GBSS expression. However, total phosphate content was reduced by ~25% in two lines in which SBEII expression was increased, with both C6- and C3-linked phosphate content being reduced.
Table 2.
Phosphate content of tuber starches from lines exhibiting silencing of GBSS (1041 lines) or overexpressing SBEII (1047 lines)
| Line | C6 PO 4 | C3 PO 4 | Total |
|---|---|---|---|
| WT898 | 32.8 ± 1.8 | 7.5 ± 0.6 | 40.3 |
| EV33-5 | 29.0 ± 2.7 | 6.4 ± 0.6 | 35.4 |
| 1041-3 | 35.2 ± 6.1 | 8.8 ± 1.9 | 44.0 |
| 1041-16 | 31.7 ± 1.6 | 7.1 ± 0.5 | 38.8 |
| 1047-15 | 24.1 ± 2.5 | 5.1 ± 0.3 | 29.1 |
| 1047-17 | 24.5 ± 2.1 | 5.8 ± 0.1 | 30.2 |
WT: wild-type; EV: empty vector. Data are mean nmol Glc6P per mg starch ± SD and mean nmol Glc3P per mg starch ± SD (n = 3 biological replicates). Total nmol phosphate per mg starch was calculated as the sum of C6 plus C3 phosphates.
Effects of altered GBSS and SBEII expression on the morphology of starch granules
To test whether the morphology of the starch granules was altered by the transgenic modifications, a range of microscopy techniques was employed (Figure 5). Brightfield microscopy showed a wide range of granule sizes in wild-type starch (Figure 5A), and neither knockdown of GBSS nor overexpression of SBEII had any obvious effect on the size range distribution of the granules. Granules from wild-type and SBEII overexpressors stained dark blue with iodine (Figure 5B), whereas granules from GBSS knockdown plants showed the pale purple colour typical of a low amylose content but with visible dark blue staining at the hilum core. Examination under polarised light detected birefringence visible as a ‘Maltese cross’ shape and blue and yellow sectors (Figure 5C). Birefringence was higher in the GBSS knockdown line, presumably since these lines are higher in amylopectin content, which has a more ordered orientation (crystallinity) than amylose. An increased birefringence in the GBSS knockdown line was also observed using differential interference contrast microscopy (Figure 5D). Variable pressure scanning electron microscopy showed that the external morphology of the starch granules was not affected by GBSS knockdown or by SBEII overexpression (Figure 5E).
Figure 5.
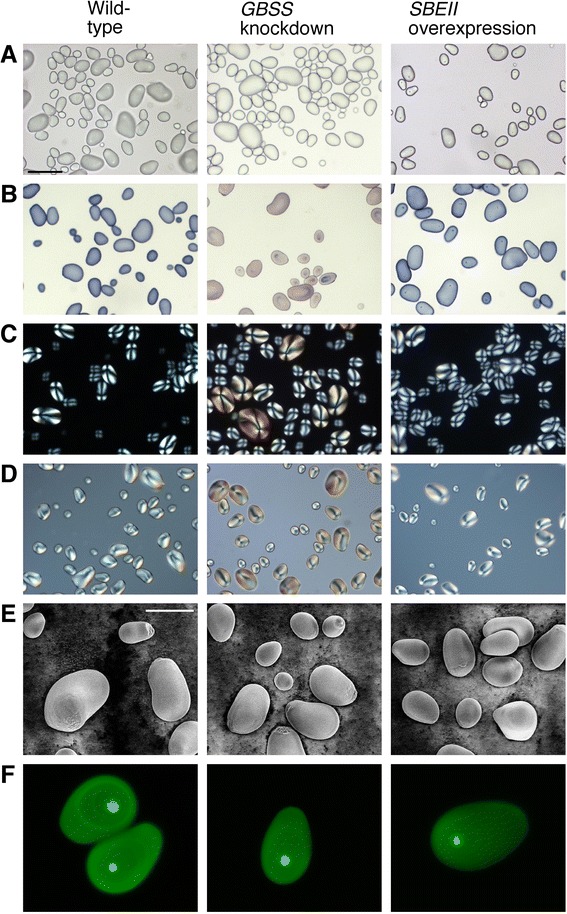
Images of tuber starch granules from wild-type and transgenically modified potato tubers. (A) Brightfield. (B) Brightfield, starch stained with I2/KI. (C) Polarised light. (D) Differential interference contrast. (E) Variable pressure scanning electron microscopy. (F) Optical section of starch fluorescently labelled with APTS taken using confocal scanning laser microscopy. Lines used were WT898, 1041–3 and 1047–17. The scale bar (in panel A) for light microscopy pictures (A–D) represents 100 μm, and for the scanning electron microscopy (E) represents 40 μm.
Details of the internal structure of the granules were examined by fluorescent labelling with 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) combined with confocal laser scanning microscopy (Figure 5F). APTS labels the single reducing end of each molecule with high efficiency, and since amylose molecules are much smaller than the highly branched amylopectin molecules, they exhibit a higher fluorescence (on a fluorescently-labelled reducing end per total Glc residue basis). A small intensely labelled region at the hilum core was visible in all the lines, indicating a high concentration of amylose at this site. This was observed even in the low amylose GBSS knockdown line, and is consistent with the iodine staining of this site seen under brightfield microscopy. Growth rings were more visible in the wild-type, suggesting that the deposition of amorphous and crystalline growth rings may have been altered in the transgenic lines. However, overall fluorescence was lower in both of the transgenic lines, and was too low for clear visualisation of growth rings in the SBEII overexpressors.
Thermal and gelatinisation properties of transgenically modified starch
The different contents of amylose, short-branched amylopectin and phosphorus in the starch would be expected to result in altered thermal and gelatinisation properties. Both knockdown of GBSS expression and overexpression of SBEII markedly altered the onset of gelatinisation temperature (To), but in opposite ways (Table 3). In wild-type and empty vector controls, To ranged from 54.2 to 55.4°C, whereas in three lines with reduced GBSS expression and lowered amylose content (1041–1, 3 and 16), To was increased by ~4°C, to 59.3 to 60.0°C. In the transgenic line 1041–4, with unaltered GBSS expression, To was not increased relative to controls. In contrast, overexpression of SBEII in various 1047 lines reduced To by up to 3°C (Table 3). In these lines, a trend towards lowered To (WT > 1047–5 = 1047–23 > 1047–12 ≥ 1047–15 ≥ 1047–17) correlated strongly (r = −0.931) with the trend of increasing SBEII activity (Figure 2). In line 1047–3, which showed SBEII activity similar to wild-type, To was not reduced and was within the range of variability seen in control lines. The temperature of gelatinisation peak (Tp), temperature of gelatinisation conclusion (Tc) and gelatinisation enthalpy (ΔH) showed similar trends to To in all of the lines.
Table 3.
Thermal properties of tuber starch granules from plants exhibiting silencing of GBSS (1041 lines) or overexpressing SBEII (1047 lines)
| Line | T o (°C) | T p (°C) | T c (°C) | Δ H (J g −1 ) |
|---|---|---|---|---|
| WT676 | 54.7 ± 0.7e | 58.1 ± 0.4ef | 61.6 ± 0.0d | 15.4 ± 0.4bcde |
| WT898 | 55.3 ± 0.2e | 58.5 ± 0.1ef | 62.1 ± 0.1d | 15.9 ± 0.2bcde |
| WT903 | 55.4 ± 0.6e | 58.6 ± 0.3ef | 61.6 ± 0.0d | 15.7 ± 0.1bcde |
| EV33-3 | 54.2 ± 0.7cd | 57.7 ± 0.4d | 61.6 ± 0.5d | 15.8 ± 2.7bcde |
| EV33-5 | 55.4 ± 0.6e | 58.7 ± 0.4fg | 62.1 ± 0.1d | 15.0 ± 0.5abcd |
| EV34-33 | 54.9 ± 0.2de | 58.2 ± 0.1de | 61.7 ± 0.0d | 14.9 ± 0.5abc |
| 1041-1 | 59.3 ± 0.1f | 63.1 ± 0.2h | 67.5 ± 0.1f | 18.2 ± 0.1f |
| 1041-3 | 59.6 ± 0.6f | 63.8 ± 0.4i | 68.7 ± 0.3g | 17.2 ± 1.5ef |
| 1041-4 | 53.8 ± 0.5c | 57.8 ± 0.2d | 61.9 ± 0.6d | 16.8 ± 1.7def |
| 1041-16 | 60.0 ± 0.4f | 63.4 ± 0.3hi | 67.9 ± 0.3f | 17.9 ± 0.4f |
| 1047-3 | 55.2 ± 0.3e | 59.1 ± 0.5g | 64.9 ± 1.2e | 16.6 ± 1.5cdef |
| 1047-5 | 53.6 ± 0.8c | 57.0 ± 0.7c | 60.9 ± 0.1c | 14.2 ± 1.6ab |
| 1047-12 | 52.8 ± 0.9b | 55.9 ± 0.5b | 59.1 ± 0.2ab | 13.7 ± 0.5a |
| 1047-15 | 52.3 ± 0.4ab | 55.6 ± 0.2b | 59.2 ± 0.2b | 14.5 ± 0.5ab |
| 1047-17 | 52.0 ± 0.3a | 55.0 ± 0.1a | 58.5 ± 0.2a | 14.6 ± 0.4ab |
| 1047-23 | 53.7 ± 0.2c | 56.7 ± 0.2c | 59.7 ± 0.3b | 15.0 ± 1.3abcd |
WT: wild-type; EV: empty vector. Data are means ± SD (n = 4 biological replicates). Means in the same column not sharing a common letter are significantly different between groups at P = 0.05 as determined by LSD after a one-way ANOVA test.
T o = temperature of onset of gelatinisation peak.
T p = temperature of gelatinisation peak.
T c = temperature of gelatinisation conclusion.
ΔH = gelatinisation enthalpy of starch.
An examination of starch pasting properties using a rapid visco analyser (RVA) detected differences between the lines (Figure 6), including the lowered To of starch from line 1047–17 and the increased To of starch from line 1041–3. Other than the onset temperature, pasting profiles were similar in shape, but with a reduction in peak viscosity of ~10% for line 1047–17 and ~24% for line 1041–3. Final viscosities were also reduced in both transgenic lines.
Figure 6.
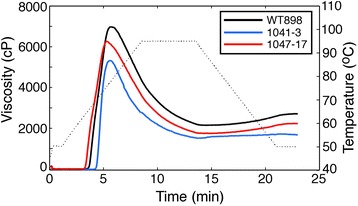
Comparison of pasting properties of tuber starch from wild-type and transgenic lines. Pasting properties were assessed in starch from lines exhibiting GBSS knockdown (1041–3) or SBEII overexpression (1047–17). Changes in viscosity of 7% (w/w) starch suspensions during heating and subsequent cooling were measured by RVA. Curves are the means of measurements on three independent biological replicates per line. The thin dotted line shows the change in temperature during measurement.
The greatest difference in physicochemical properties was seen in the gelatinisation of starch granules in urea. In 4 M urea, starch granules from line 1041–3 showed very little gelatinisation, with a swollen pellet volume only one-third that of wild-type starch granules (Figure 7A). Starch from lines 1041–1 and 1041–16 produced similar results (data not shown). In contrast, starch granules from line 1047–17 exhibited a huge amount of gelatinisation, producing a large and diffuse pellet with a swollen volume 3-fold greater than that of wild-type. Starch from line 1047–15 produced similar results (data not shown). Examination of a range of urea concentrations showed that gelatinisation was initiated at a lower urea concentration in starch granules from 1047–17, but followed a similar curve to wild-type and achieved a similar maximum swollen volume (Figure 7B). Gelatinisation followed a very different pattern in starch granules from 1041–3, beginning at a higher urea concentration and showing a remarkably steep increase in swollen pellet volume over a change in urea concentration from 4 M to 6 M. The maximum swelling volume achieved by 1041–3 was up to 25% greater than wild-type, and at urea concentrations above 7 M occupied the entire 1 mL volume of the original liquid provided.
Figure 7.
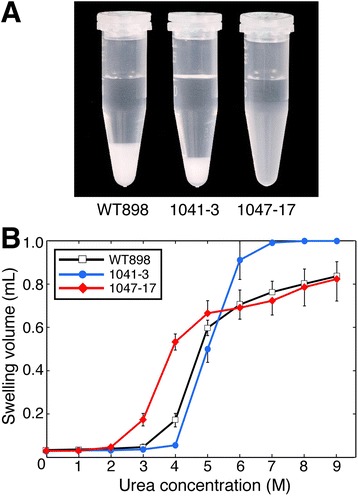
Changes in gelatinisation of tuber starch granules from transgenic potato lines. (A) Effect of GBSS knockdown (1041–3) or SBEII overexpression (1047–17) on starch granule swelling in 4 M urea. (B) Comparison of swelling of starch granules in a range of urea concentrations. Gelatinisation was assessed by measuring the volume of the swollen starch pellet. Data are mean pellet volumes ± SD (n = 4 biological replicates).
The extent of SBEII overexpression had a clear effect on the swollen volume of the starch granules in 4 M urea (Figure 8). In line 1047–3, swollen pellet volume was less than wild-type. However, the other lines formed a series of increasing swollen pellet volume, in the order WT < 1047–5 = 1047–23 < 1047–12 ≤ 1047–17 ≤ 1047–15. This order reflected the trend in increasing SBEII activity (Figure 2), confirming that SBEII activity and starch swelling power are positively correlated (r = 0.939). In contrast, the increase in swollen pellet volume was strongly but negatively correlated (r = −0.976) to the reduction of To (Table 3).
Figure 8.
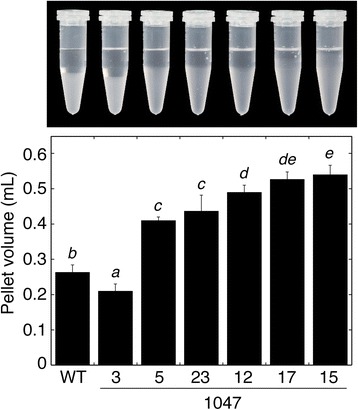
Effects of altered SBEII expression on the gelatinisation of tuber starch granules in 4 M urea. Data are mean pellet volumes ± SD (n = 3 biological replicates). Means not sharing a common letter are significantly different between groups at P = 0.05 as determined by LSD after a one-way ANOVA test. A photograph of one of the replicates is shown above the histogram.
Discussion
The manipulation of starch properties is important commercially for various industrial applications, as well as for the production of food ingredients. Several loss-of-function mutants and RNAi knockdown studies have helped elucidate the role of particular enzymes in starch biosynthesis [3], but overexpression of starch biosynthetic enzymes is more difficult to accomplish and few examples exist. In one study, overexpression of SBEIIb in rice produced starch with increased branching, reduced To, increased solubility and increased swelling power [22]. However, potato starch has several advantages over cereal starch for commercial applications. Rice starch granules are small (around 5 μm) and polygonal with sharp edges [29], a high lipid content and a high To, whereas potato starch granules are of a large oval shape of average size 40 μm and are less crystalline than rice starch [8,30,31]. Starch from potato is also highly phosphorylated relative to cereal starch [32]. The large granule size, low crystallinity, low lipid content and high phosphate content give potato starch a low temperature for onset of gelatinisation, and a high swelling power and viscosity that are useful for both culinary and industrial applications.
Amylopectin chains have a polymodal size distribution, and models of amylopectin structure propose a long C chain that contains the single reducing terminal residue, long inner B chains that carry additional branches, and shorter, outer A chains that do not carry additional branches and are linked to the B chains [33]. Polymodal chain size distributions of DP 6–12, 13–24, 25–36 and ≥37 are believed to correspond to A chains (DP6–12), and various classes of longer B chains [34]. The abundance of short A chains of DP 6–12 probably determines the starch crystalline allomorph [31]. A range of SBEII overexpression levels increased the number of amylopectin chains of DP ≤ 12 and caused a particularly large increase in those of DP6 (Figure 4B). This did not result in any obvious difference in the shape or size of the granules or in their iodine staining properties (Figure 5), presumably because amylose content was not substantially affected (Table 1). Iodine staining is largely due to the content of amylose, which has a 20-fold stronger binding capacity for iodine than does amylopectin [26]. In contrast, knockdown of GBSS resulted in a pale purple colour upon iodine staining (Figure 5B), indicating a low amylose content, but with dark blue staining and intense fluorescence at the hilum (Figure 5B,F). A high concentration of amylose at the hilum core even in GBSS-silenced lines is consistent with previous observations [6,27]. Knockdown of GBSS did not bring about an alteration in the appearance of the granules (Figure 5), [27].
Starch gelatinisation, caused by heating starch in excess water (or exposure to chaotropic agents such as urea), is due to disruption of the molecular order in the crystalline regions and results in irreversible granular swelling and starch solubilisation. The temperature required for starch gelatinisation, and the swelling volume achieved, are affected by granule size and shape, the amylose:amylopectin ratio, amylopectin branching, phosphorus content and other factors. Overexpression of SBEII created amylopectin with a high abundance of short DP6 side chains and facilitated gelatinisation, enabling gelatinisation to occur at a lower temperature (Table 3) and at a lower concentration of urea (Figure 7). Granule size and shape were not affected by GBSS knockdown or SBEII overexpression, but amylose content, amylopectin branching and phosphorus content were altered. Starch phosphorylation occurs mainly on the longer branches of amylopectin, with only a minor proportion on short amylopectin chains, and is essentially absent on amylose [28,35]. Thus, knockdown of GBSS expression and the resulting low amylose content had little effect on the degree of starch phosphorylation (Table 2). In SBEII overexpression lines, the increased number of short branches of DP ≤ 12 and reduced number of longer branches of DP ≥ 20 should reduce total starch phosphorus content to some extent, and a 25% reduction was observed.
Studies on cereal starch have suggested that swelling power is exclusively due to the properties of the amylopectin, with amylose and lipids inhibiting swelling [36]. In SBEII-overexpressing line 1047–17, the swelling volume in 4 M urea was increased to approximately 65% of the maximal volume achieved (in 9 M urea), compared with only 20% for wild-type (Figure 7). A very low amylose content in line 1041–3 changed the gelatinisation response to urea concentration, and increased swelling power at >6 M urea (Figure 7), [17,37], but amylose content in the SBEII overexpressing lines was hardly different from wild-type (Table 1). This suggests that amylopectin branching pattern was the major factor influencing starch swelling power. Previous studies have found that To decreased when there was an increased frequency of short amylopectin branches, due either to knockdown of isoamylase1 or overexpression of SBEIIb in rice [22,29], or knockdown of SSII/SSIII or overexpression of bacterial glycogen branching enzyme in potato [21,38]. The converse finding, that an increased Tp results from a reduction in the frequency of short branches due to knockdown of SBEII genes in rice and barley [9,10] or the amylose extender (knockout of SBEIIb) mutant of maize [7], confirms the influence of starch branching frequency on thermal properties.
In lines with a range of SBEII expression, there was a correlation between SBEII activity, the degree of reduction in To and the degree of increase in the swelling volume in 4 M urea. In other studies, comparisons between transgenic potato lines found that swelling power was decreased when either amylopectin branching was reduced (knockdown of SBEI/SBEII), or phosphorus content was reduced (knockdown of GWD (glucan, water dikinase)) [39]. A knockdown of GWD that reduced starch C6-linked phosphorus content by 85% had little effect on To orTp, although swelling power was considerably reduced [39,40]. We observed increased swelling power in starch from our SBEII overexpressing lines, despite a 25% reduction in phosphorus content. Thus, increased amylopectin branching rather than small changes in amylose or phosphorus content most likely produced both the lowered To and increased swelling power. Consistent with the data presented here, increased branching in rice starch was also reported to increase swelling volume [22], although differences were not quantified and the swelling power of rice starch is very small relative to that of potato starch [41].
The present study shows that overexpression of potato SBEII increases the proportion of short chains of DP6-12, consistent with in vitro experiments that found potato SBEII transfers a high number of chains of DP6 [42] and knockdown of SBEII that caused an increase in average chain length [14]. We propose that this results in an abnormal amylopectin structure with reduced crystallinity, thus altering gelatinisation properties. In contrast, the increase in To found in the GBSS knockdown lines was most likely due to the reduced amylose content, since the degree of amylopectin branching and the phosphorus content were not substantially altered (Figure 4D, Table 2). An increase in the gelatinisation temperature of low-amylose potato starch has been noted previously [37,39].
Both genetic manipulations caused a lowered peak viscosity as determined by RVA, which measures changes in viscosity of the starch as it is heated and cooled under constant stirring (Figure 6). A rapid rise in viscosity upon heating caused by gelatinisation is followed by a decline as the amylopectin matrix structure is broken down by mechanical shearing. The two amylose-containing starches (WT898 and 1047–17) showed a higher peak viscosity and a slightly higher setback viscosity after cooling than line 1041–3. The reduced peak and final viscosities of low-amylose starches, such as that from 1041–3, are consistent with previous studies [15,43]. Starch from a line overexpressing SBEII also had a reduced viscosity, presumably due to the increased ratio of short to long amylopectin branches [21]. This is useful commercially since a reduction in the viscosity of potato starch is highly desirable in the production of manufactured products, in order to facilitate handling [37].
Conclusions
The data presented here show that overexpression of the potato SBEII gene can be achieved in potato, and that a relatively small single-intron construct obviates the difficulties of either manipulating a very large genomic fragment or cloning a functional SBEII cDNA gene in bacteria. The use of potato rather than cereals is advantageous since transformation is more facile and starch yields are high. The abundance of short amylopectin branches and thermal properties are highly correlated, and elsewhere highly branched starch with many short chains has been shown to have a lower gelatinisation temperature and improved rheological properties [44]. The To of native potato starch is up to 10°C lower than that of rice starch [41], and in the work presented here the increased abundance of short amylopectin branches has lowered it by another 3°C and reduced viscosity while increasing swelling power. Such starches may be expected to find many uses in the food and manufacturing industries.
Additionally, although modifying the amylopectin branching pattern in planta requires the use of genetic modification, starch produced in this way may replace chemically modified starch in food applications and thus remove environmental concerns and food additive labelling requirements. Previously, studies of overexpression of starch branching enzymes in potato have used bacterial enzymes [19-21]. Since the transgene used for overexpression in the current work consisted of a modified potato SBEII gene that did not contain any non-potato elements and was controlled by a potato promoter and terminator (both from the GBSS gene), the approach could be used to create entirely intragenic plants where all the transferred DNA (including T-DNA borders) is derived from the potato genome [45]. The use of intragenic technologies with all-potato DNA may ultimately remove the need for genetic modification labelling [45].
Methods
Construct assembly and transgenic plants
It did not prove possible to propagate a complete cDNA of potato SBEII in Escherichia coli, so an alternative strategy was employed. A suitable short intron to prevent translation of a functional SBEII protein in bacteria was identified at amino acid 439, and primers (Additional file 1: Table S1) were designed to incorporate existing AflII and SacI sites in the potato sequence that flanked the putative intron. PCR amplification from potato (cv ‘Karaka’) gDNA using primers FW3 and RW10 produced a 374 bp fragment that contained an intron of 231 bp.
The upstream portion of potato SBEII cDNA was PCR-amplified using primers SBE-A and RW11 (incorporating a AflII site) to give cDNA1, and the downstream portion using primers F4 (incorporating a SacI site) and SBE-B to give cDNA2. All PCR products were verified by DNA sequencing. cDNA1 and the genomic fragment containing the intron were ligated together using the AflII site, then the cDNA1:gDNA fragment (blunt-SacI) and the cDNA2 fragment (SacI-blunt) were ligated in a three-way ligation into expression vector GBSSPTpUC57 (a gift from Dr A. J. Conner), between the GBSS promoter and the GBSS terminator. This produced a 4.46 kb construct pDB1047 consisting of GBSSpro:cDNA1:gDNA:cDNA2:GBSSterm (Additional file 1: Figure S1).
To produce an inverted repeat construct for silencing of GBSS, two fragments of potato GBSS cDNA were amplified by PCR using primer pairs GBSS-A and GBSS-B, and GBSS-C and GBSS-D (Additional file 1: Table S1). Primers GBSS-A and GBSS-B amplified a fragment of 1364 bp spanning amino acids 132–585 of the 607-amino acid protein, adding an initiating methionine codon and restriction sites XhoI upstream and PstI downstream. Primers GBSS-C and GBSS-D amplified a fragment of 987 bp spanning amino acids 135–463, adding restriction sites XbaI and StuI upstream and PstI downstream. The GBSS-A/GBSS-B PCR product was ligated in the sense orientation to the GBSS-C/GBSS-D PCR product in the antisense orientation, using the PstI site. This DNA was ligated into expression vector GBSSPTpUC57 between the GBSS promoter and the GBSS terminator to produce construct pJZ1041.
Construct pDB1047 was transferred into binary vector pMOA33, conferring resistance to kanamycin, and construct pJZ1041 was transferred into binary vector pMOA34, conferring resistance to hygromycin [46]. These binary vectors were electroporated into Agrobacterium tumefaciens LBA4404, which was used to transform Solanum tuberosum cv ‘Karaka’, a cultivar common in New Zealand, as described [47]. Plantlets were confirmed as transgenic by screening for the presence of the transgene and the selectable marker gene using PCR (Additional file 1: Figure S2). Empty vector transformants were produced similarly, and non-transformed wild-type (WT) material was regenerated through the tissue culture process (lacking the selectable antibiotic) as controls. Primary transformants were grown and screened as described below.
Plant growth and molecular analysis
Plantlets were potted in plastic bags (16 L) of potting mix and grown to maturity in a containment glasshouse in Palmerston North (40° 21′ S, 175° 36′ E) during the southern summer and autumn months (January to May). Glasshouse conditions were approximately 15 h daylight per day, with supplementary cooling and heating providing a maximum daytime temperature of 27°C and a minimum night-time temperature of 16°C. The plant aerial portions were allowed to senesce naturally. Three tubers per plant were peeled, chopped into small cubes and frozen in liquid nitrogen before being stored at −80°C for RNA or enzyme assay. RNA was prepared using the hot borate method [48], and 10 μg per lane was separated by electrophoresis in 1.2% agarose denaturing formaldehyde gels. The RNA was blotted to Hybond-XL nylon membrane (GE Healthcare, Little Chalfont, UK) and immobilized by ultraviolet irradiation. Labelled probes were produced using PCR products SBEII cDNA2 and GBSS-A/GBSS-B as templates with [α-32P]dATP and the Klenow fragment of DNA polymerase I. RNA gel blots were hybridised overnight with the labelled probes in Church and Gilbert [49] buffer at 65°C, washed in 1x SSC at 65°C and exposed to X-ray film. For analysis of starch physicochemical properties, independent primary transformants were multiplied by excising axillary buds of plantlets in tissue culture and regenerating them to whole plants and tubers in subsequent summers. Biological replicates consisted of pooled tissue of three tubers from three or four different but genetically identical plants.
Native PAGE determination of SBEII activity
Tuber tissue was powdered in liquid nitrogen, and 100 mg was extracted with 100 μL of extraction buffer (50 mM HEPES-NaOH, pH 7.4, 2 mM MgCl2, 50 mM 2-mercaptoethanol, 12.5% (v/v) glycerol), incubated on ice for 5 min then centrifuged twice at 16,000 × g for 15 min [17]. Soluble proteins in the supernatant were used as the crude enzyme preparation. Proteins (150 μg per lane) were separated on native 5% (w/v) polyacrylamide gels containing substrates for SBE activity as described by Tetlow et al. [24]. After electrophoresis, gels were rinsed, incubated in reaction buffer at 30°C for 2 h then stained with I2/KI as described by Nishi et al. [17]. Gels were photographed immediately after staining. Gel pictures were converted to greyscale, and relative density in the SBEII spots quantified using MultiGauge software (Fujifilm, Tokyo, Japan).
Starch preparation
Starch was prepared by chopping peeled fresh tubers into small pieces, adding an equal amount of ice-cold water and crushing the tissue with a potato masher. Starch granules were washed through a 200 μm mesh sieve, and the suspension allowed to settle overnight. The supernatant and any discoloured layer above the white starch pellet were discarded. The starch was resuspended in a large volume of ice-cold water and the settling step repeated. The pellet of starch granules was freeze-dried and used for analysis.
Microscopy
Aqueous, unstained suspensions of starch grains were observed by brightfield, differential interference contrast (DIC) and simple polarising microscopy (crossed polars but no phase plate) using an Olympus Vanox AHT3 microscope (Olympus Optical Co Ltd., Tokyo, Japan). Brightfield microscopy was also used to view starch stained with iodine using a stain solution of 0.2% I2 in a 2% aqueous solution of KI at a dilution ratio of 1:100 stain:starch suspension. For scanning electron microscopy (SEM), dry starch powder was lightly sprinkled onto carbon adhesive tabs attached to aluminium stubs and was viewed uncoated in a FEI Quanta 250 SEM (FEI Company, Hillsboro, OR, USA) using variable pressure mode to reduce charging (chamber pressure 60 Pa, accelerating voltage 5 kV).
The reducing ends of starch molecules were fluorescently labelled within the starch granules using APTS as described by Blennow et al. [27]. Optical sections were taken under a Leica SP5 DM6000B laser scanning confocal microscope, using an excitation wavelength of 488 nm and detection at the interval from 500 to 535 nm.
Size exclusion chromatography
Aliquots of starch were suspended in 2 M NaOH, gelatinised at 100°C then dissolved in water as described by Blennow et al. [26]. The solubilised starch was applied to a Waters Ultrahydrogel Linear column (300 × 7.8 mm) with an exclusion limit of 7 × 106 Da, and eluted with 10 mM NaOH at 50°C. Carbohydrate content in the effluent was recorded on-line by measuring refractive index (Waters 2414 Refractive Index detector (Waters Corp, MA, USA)). A large peak at the end of the elution profiles (due to NaOH) was omitted.
Starch composition
The percentage of amylose present in the starch was measured using the Megazyme K_AMYL Amylose kit (Megazyme, Bray, Ireland). Starch branching patterns were assessed using a micro-scale FACE procedure to measure chain length size distribution [50]. The degree of C6 and C3 starch phosphorylation was determined using a mass spectrometric method [51].
Starch properties
Thermal analysis of gelatinisation properties was performed using a Q2000 differential scanning calorimeter (DSC) (TA Instruments, New Castle, DE, USA). Starch (~5 mg) was accurately weighed into Tzero aluminium hermetic pans (TA Instruments), and 2.33 μL water mg−1 starch added to give a suspension containing 70% water. The pans were hermetically sealed and left at room temperature (20°C) for at least 2 h. DSC conditions were: equilibration at 10°C for 5 min, then heating at 5°C min−1 to 100°C. The heat flow curves were analysed using TA Universal Analysis 2000 software version 4.5A build 4.5.0.5 using the “integrate peak sigmoid horizontal” option over the range 50–80°C. Variables calculated were peak onset temperature (To), peak maximum temperature (Tp), peak conclusion temperature (Tc) and peak area (ΔH), which provides an estimate of enthalpy in J g−1.
Starch pasting properties were evaluated using a rapid visco analyser (RVA) (Newport Scientific, Sydney, Australia). Approximately 1 g of starch was accurately weighed and dried at 130°C to constant weight. A starch suspension was prepared at 7% (w/w) on a dry weight basis in distilled water, with a total weight of 28 g. The suspension was equilibrated at 50°C for 1 min, heated to 95°C at 6°C min−1, maintained at 95°C for 5 min, then cooled to 50°C at 6°C min−1, with constant stirring at 160 rpm.
The swelling volume of starch was determined using a urea procedure [17]. Freeze-dried starch (20 mg) was mixed with 1 mL of urea solution (0 to 9 M). The mixtures were allowed to stand at room temperature (23°C) for 24 h, centrifuged at 8000 × g for 20 min, then allowed to stand at room temperature for 1 h. The volumes of the swollen pellet were determined by subtracting the volume of the supernatant from the original 1 mL volume.
Statistical analysis
Means were compared using one-way ANOVA. Least significant differences (LSDs) were used to establish which means differed significantly (P = 0.05). Since for SBEII activity the data was relative and was scaled so that wild-type was 1, the wild-type data was not included in the ANOVA; pooled standard errors from the ANOVA were used in one-sample t-tests to test whether each mean was significantly different from 1. Correlation coefficients were calculated between means. Analysis was carried out using Genstat (version 17, 2014, VSNI Ltd, Hemel Hempstead, UK).
Acknowledgements
We thank Tony Conner for the generous gift of GBSSPTpUC57 plasmid, Jianyu Chen (Manawatu Microscopy and Imaging Centre, Massey University) for assistance with confocal microscopy, Kerry Sullivan and Ronan Chen for assistance with RNA preparation, Tonya Frew and Rebecca Cooper for running the FACE analysis, Nigel Joyce for mass spectrometer operations, Ian King for care of the plants, Duncan Hedderley for statistical analysis and Nigel Larsen and Paul Johnston for comments on the manuscript. This work was funded by contract number C02X0805 from the Foundation for Research, Science and Technology of New Zealand.
Abbreviations
- APTS
8-amino-1,3,6-pyrenetrisulfonic acid
- DP
Degree of polymerisation
- DSC
Differential scanning calorimeter
- FACE
Fluorophore-assisted carbohydrate electrophoresis
- GBSS
Granule-bound starch synthase
- PAGE
Polyacrylamide gel electrophoresis
- RVA
Rapid visco analyser
- SBE
Starch branching enzyme
- SS
Starch synthase
Additional file
Primers used for PCR amplification. Figure S1. Sequence of the hybrid cDNA/gDNA SBEII-intron construct. Figure S2. PCR screening of transgenic plants for the presence of the transgene and the selectable marker gene. Figure S3. RNA gel blot analysis of SSII and SSIII mRNA abundance in tubers from plants overexpressing SBEII.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DAB conceived and designed the study, DAB, LWM, JZ, MJM, ICH, LS, MC and GMT-V performed experiments, DAB wrote the paper. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Contributor Information
David A Brummell, Email: david.brummell@plantandfood.co.nz.
Lyn M Watson, Email: lynwatsonnz@gmail.com.
Jun Zhou, Email: jun_zcn@hotmail.com.
Marian J McKenzie, Email: marian.mckenzie@plantandfood.co.nz.
Ian C Hallett, Email: ian.hallett@plantandfood.co.nz.
Lyall Simmons, Email: lyall.simmons@plantandfood.co.nz.
Margaret Carpenter, Email: margaret.carpenter@plantandfood.co.nz.
Gail M Timmerman-Vaughan, Email: gail.timmerman-vaughan@plantandfood.co.nz.
References
- 1.Jobling S. Improving starch for food and industrial applications. Curr Opinion Plant Biol. 2004;7:210–8. doi: 10.1016/j.pbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209–34. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 3.Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol. 2003;54:207–33. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 4.Kötting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment? Curr Opinion Plant Biol. 2010;13:321–9. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Yoo SH, Jane J. Structural and physical characteristics of waxy and other wheat starches. Carbohydr Poly. 2002;49:297–305. doi: 10.1016/S0144-8617(01)00338-1. [DOI] [Google Scholar]
- 6.Visser RGF, Suurs LCJM, Bruinenberg PM, Bleeker I, Jacobsen E. Comparison between amylose-free and amylose containing potatoes. Starch-Stärke. 1997;49:438–43. doi: 10.1002/star.19970491103. [DOI] [Google Scholar]
- 7.Yuan RC, Thompson DB, Boyer CD. Fine structure of amylopectin in relation to gelatinization and retrogradation behavior of maize starches from three wx-containing genotypes in two inbred lines. Cereal Chem. 1993;70:81–9. [Google Scholar]
- 8.Srichuwong S, Sunarti TC, Mishima T, Isono N, Hisamatsu M. Starches from different botanical sources I: Contribution of amylopectin fine structure to thermal properties and enzyme digestibility. Carbohydr Poly. 2005;60:529–38. doi: 10.1016/j.carbpol.2005.03.004. [DOI] [Google Scholar]
- 9.Butardo VM, Fitzgerald MA, Bird AR, Gidley MJ, Flanagan BM, Larroque O. et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J Exp Bot. 2011;62:4927–41. [DOI] [PMC free article] [PubMed]
- 10.Regina A, Kosar-Hashemi B, Ling S, Li Z, Rahman S, Morell M. Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J Exp Bot. 2010;61:1469–82. doi: 10.1093/jxb/erq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carciofi M, Blennow A, Jensen SL, Shaik SS, Henriksen A, Buléon A, et al. Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol. 2012;12:223. doi: 10.1186/1471-2229-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura Y, Utsumi Y, Sawada T, Aihara S, Utsumi C, Yoshida M, et al. Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol. 2010;51:776–94. doi: 10.1093/pcp/pcq035. [DOI] [PubMed] [Google Scholar]
- 13.Larsson CT, Khoshnoodi J, Ek B, Rask L, Larsson H. Molecular cloning and characterization of starch-branching enzyme II from potato. Plant Mol Biol. 1998;37:505–11. doi: 10.1023/A:1005908305456. [DOI] [PubMed] [Google Scholar]
- 14.Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, et al. A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J. 1999;18:163–71. doi: 10.1046/j.1365-313X.1999.00441.x. [DOI] [PubMed] [Google Scholar]
- 15.Flipse E, Suurs LCJM, Keetels CJAM, Kossmann J, Jacobsen E, Visser RGF. Introduction of sense and antisense cDNA for branching enzyme in the amylose-free potato mutant leads to physico-chemical changes in the starch. Planta. 1996;198:340–7. doi: 10.1007/BF00620049. [DOI] [Google Scholar]
- 16.Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, et al. Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydr Poly. 1998;35:155–68. doi: 10.1016/S0144-8617(97)00249-X. [DOI] [Google Scholar]
- 17.Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of Amylose-Extender mutation in rice endosperm. Plant Physiol. 2001;127:459–72. doi: 10.1104/pp.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, et al. Production of very-high-amylose potato starch by inhibition of SBE A and B. Nature Biotechnol. 2000;18:551–4. doi: 10.1038/75427. [DOI] [PubMed] [Google Scholar]
- 19.Shewmaker CK, Boyer CD, Wiesenborn DP, Thompson DB, Boersig MR, Oakes JV, et al. Expression of Escherichia coli glycogen synthase in the tubers of transgenic potatoes (Solanum tuberosum) results in a highly branched starch. Plant Physiol. 1994;104:1159–66. doi: 10.1104/pp.104.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortstee AJ, Vermeesch AMS, de Vries BJ, Jacobsen E, Visser RGF. Expression of Escherichia coli branching enzyme in tubers of amylose-free transgenic potato leads to an increased branching degree of the amylopectin. Plant J. 1996;10:83–90. doi: 10.1046/j.1365-313X.1996.10010083.x. [DOI] [PubMed] [Google Scholar]
- 21.Kortstee AJ, Suurs LCJM, Vermeesch AMG, Keetels CJAM, Jacobsen E, Visser RGF. The influence of an increased degree of branching on the physico-chemical properties of starch from genetically modified potato. Carbohydr Poly. 1998;37:173–84. doi: 10.1016/S0144-8617(98)00045-9. [DOI] [Google Scholar]
- 22.Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, et al. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol J. 2004;2:507–16. doi: 10.1111/j.1467-7652.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- 23.FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Division. 2013. http://faostat3.fao.org. Accessed 19 February 2015.
- 24.Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, et al. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–409. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blennow A, Bay-Smidt AM, Bauer R. Amylopectin aggregation as a function of starch phosphate content studied by size exclusion chromatography and on-line refractive index and light scattering. Int J Biol Macromol. 2001;28:409–20. doi: 10.1016/S0141-8130(01)00133-7. [DOI] [PubMed] [Google Scholar]
- 27.Blennow A, Hansen M, Schulz A, Jørgensen K, Donald AM, Sanderson J. The molecular deposition of transgenically modified starch in the starch granule as imaged by functional microscopy. J Struct Biol. 2003;143:229–41. doi: 10.1016/j.jsb.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Blennow A, Bay-Smidt AM, Wischmann B, Olsen CE, Møller BL. The degree of starch phosphorylation is related to the chain length distribution of the neutral and the phosphorylated chains of amylopectin. Carbohydr Res. 1998;307:45–54. doi: 10.1016/S0008-6215(98)00015-9. [DOI] [Google Scholar]
- 29.Fujita N, Kubo A, Suh DS, Wong KS, Jane JL, Ozawa K, et al. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol. 2003;44:607–18. doi: 10.1093/pcp/pcg079. [DOI] [PubMed] [Google Scholar]
- 30.Hizukuri S, Kaneko T, Takeda Y. Measurement of the chain length of amylopectin and its relevance to the origin of crystalline polymorphism of starch granules. Biochim Biophys Acta. 1983;760:188–91. doi: 10.1016/0304-4165(83)90142-3. [DOI] [Google Scholar]
- 31.Buléon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/S0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 32.Lim ST, Kasemsuwan T, Jane JL. Characterization of phosphorus in starch by 31P-nuclear magnetic resonance spectroscopy. Cereal Chem. 1994;71:488–93. [Google Scholar]
- 33.Peat S, Whelan WJ, Thomas GJ. Evidence of multiple branching in waxy maize starch. J Chem Soc. 1952;1952:4546–8. [Google Scholar]
- 34.Hanashiro I, Abe JI, Hizukuri S. A periodic distribution of the chain length of amylopectin as revealed by high-performance anion-exchange chromatography. Carbohydr Res. 1996;283:151–9. doi: 10.1016/0008-6215(95)00408-4. [DOI] [Google Scholar]
- 35.Takeda Y, Hizukuri S. Location of phosphate groups in potato amylopectin. Carbohydr Res. 1982;102:321–7. doi: 10.1016/S0008-6215(00)88077-5. [DOI] [Google Scholar]
- 36.Tester RF, Morrison WR. Swelling and gelatinization of cereal starches. I. Effects of amylopectin, amylose, and lipids. Cereal Chem. 1990;67:551–7. [Google Scholar]
- 37.Visser RGF, Suurs LCJM, Steeneken PAM, Jacobsen E. Some physicochemical properties of amylose-free potato starch. Starch-Stärke. 1997;49:443–8. doi: 10.1002/star.19970491104. [DOI] [Google Scholar]
- 38.Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rössner U, et al. A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 1999;17:251–61. doi: 10.1046/j.1365-313X.1999.00371.x. [DOI] [Google Scholar]
- 39.Wickramasinghe HAM, Blennow A, Noda T. Physico-chemical and degradative properties of in-planta re-structured potato starch. Carbohydr Poly. 2009;77:118–24. doi: 10.1016/j.carbpol.2008.12.013. [DOI] [Google Scholar]
- 40.Kozlov SS, Blennow A, Krivandin AV, Yuryev VP. Structural and thermodynamic properties of starches extracted from GBSS and GWD suppressed potato lines. Int J Biol Macromol. 2007;40:449–60. doi: 10.1016/j.ijbiomac.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Singh N, Singh J, Kaur L, Sodhi NS, Gill BS. Morphological, thermal and rheological properties of starches from different botanical sources. Food Chem. 2003;81:219–31. doi: 10.1016/S0308-8146(02)00416-8. [DOI] [Google Scholar]
- 42.Rydberg U, Andersson L, Andersson R, Aman P, Larsson H. Comparison of starch branching enzyme I and II from potato. Eur J Biochem. 2001;268:6140–5. doi: 10.1046/j.0014-2956.2001.02568.x. [DOI] [PubMed] [Google Scholar]
- 43.Jobling SA, Westcott RJ, Tayal A, Jeffcoat R, Schwall GP. Production of a freeze-thaw-stable potato starch by antisense inhibition of three starch synthase genes. Nature Biotechnol. 2002;20:295–9. doi: 10.1038/nbt0302-295. [DOI] [PubMed] [Google Scholar]
- 44.Jane J, Chen YY, Lee LF, McPherson AE, Wong KS, Radosavljevic M, et al. Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal Chem. 1999;76:629–37. doi: 10.1094/CCHEM.1999.76.5.629. [DOI] [Google Scholar]
- 45.Conner AJ, Barrell PJ, Baldwin SJ, Lokerse AS, Cooper PA, Erasmuson AK, et al. Intragenic vectors for gene transfer without foreign DNA. Euphytica. 2007;154:341–53. doi: 10.1007/s10681-006-9316-z. [DOI] [Google Scholar]
- 46.Barrell PJ, Conner AJ. Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. BioTechniques. 2006;41:708–10. doi: 10.2144/000112306. [DOI] [PubMed] [Google Scholar]
- 47.Barrell PJ, Shang YJ, Cooper PA, Conner AJ. Alternative selectable markers for potato transformation using minimal T-DNA vectors. Plant Cell Tiss Organ Cult. 2002;70:61–8. doi: 10.1023/A:1016013426923. [DOI] [Google Scholar]
- 48.Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 49.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–5. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray S, McKenzie M, Butler R, Baldwin S, Sutton K, Batey I, et al. Quantitative, small-scale, fluorophore-assisted carbohydrate electrophoresis implemented on a capillary electrophoresis-based DNA sequence analyzer. Anal Biochem. 2011;413:104–13. doi: 10.1016/j.ab.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Carpenter M, Joyce N, Butler R, Genet R, Timmerman-Vaughan G. A mass spectrometric method for quantifying C3 and C6 phosphorylation of starch. Anal Biochem. 2012;431:115–9. doi: 10.1016/j.ab.2012.09.004. [DOI] [PubMed] [Google Scholar]


