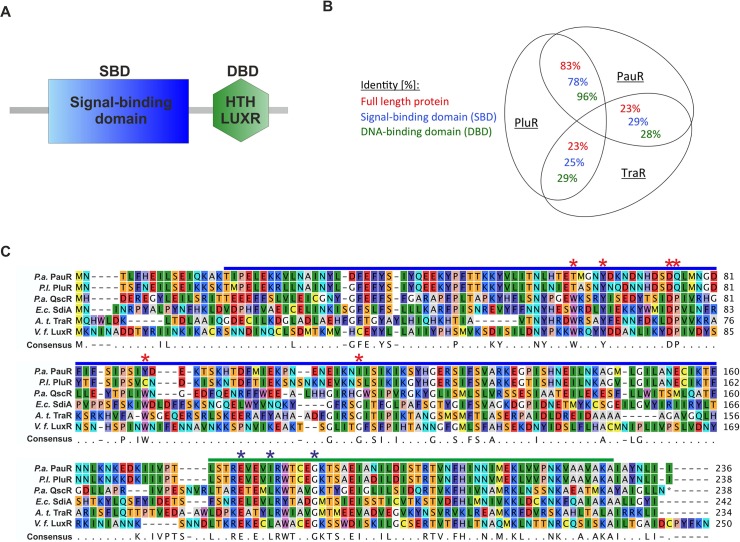
Fig 1. Protein sequence comparison of QS LuxR family members and the non-AHL sensors PluR and PauR.
(A) Modular domain structure of LuxR-type regulators, with a N-terminal signal-binding domain (SBD) and a C-terminal DNA-binding domain (DBD) [8], containing the helix-turn-helix "HTH LUXR" motif (SMART00421) [25]. (B) Comparison of the protein sequence identity of PluR from P. luminescens, PauR from P. asymbiotica and TraR from A. tumefaciens. The identity was compared either of the full-length protein sequence, only the signal-binding domain (SBD) and only the DNA-binding domain (DBD). To calculate identity of the protein sequences the LALIGN software from SIB (Swiss Institute of Bioinformatics) was used [26]. (C) Sequence alignment of the protein sequences of PauR from P. asymbiotica (P.a.), PluR from P. luminescens (P.l.), QscR from Pseudomonas aeruginosa (P.a.), SdiA from Escherichia coli (E.c.), TraR from Agrobacterium tumefaciens (A.t.) and LuxR from Vibrio fischeri (V.f.). The SBD is depicted with a blue bar and the DBD with a green bar. Within the SBD the six conserved amino acids, displaying the WYDPWG-motif of AHL-sensors, are marked with red asterisks and the three conserved amino acids in the DBD are marked with blue asterisks. Amino acids with a consensus of 60–100% are shown, positions with a lower coverage are marked with a dot. The RasMol colouring of the amino acids and the alignment was generated with CLC Mainworkbench 7 (CLC Bio Qiagen, Hilden, Germany).