Figure 4.
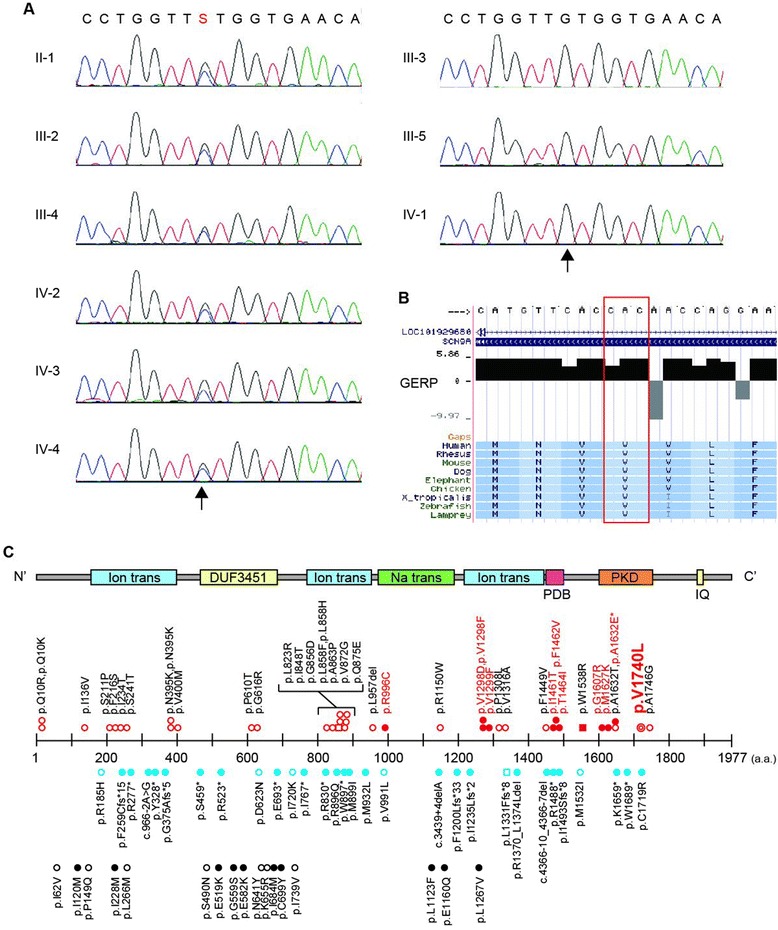
Genetic analysis of the pedigree. Electropherograms of the heterozygous SCN9A mutation. Arrow indicates the position of the mutation (NM_002977: c.5218G>C, p.Val1740Leu). Data from six affected and three unaffected individuals are shown on the left and right panels, respectively. A G/C change is represented as an “S” according to the IUPAC code. B) Evolutionary conservation of p.Val1740 in SCN9A from lampreys to humans. The altered amino acid residue is marked with a red box. The degree of evolutionary conservation was scored by Genomic Evolutionary Rate Profiling. C) The upper portion illustrates the SCN9A protein with functional domains predicted by the SMART program using the protein sequence (NP_002968). Ion trans, ion transport domain; DUF3451, DUF3451 domain; Na trans, sodium ion transport-associated domain; PDB, PDB domain; PKD, PKD (polycystic kidney) channel domain; IQ, IQ motif (calmodulin-binding motif). Middle and lower portions indicate the location of human SCN9A mutations: red double circles, mutation identified in this pedigree; red open circles, erythermalgia; red filled circles, PEPD; red open squares, pain; dysautonomia and acromesomelia; red filled squares, chronic non-paroxysmal neuropathic pain; blue open circles, small fiber neuropathy; blue open square, hereditary sensory and autonomic neuropathy type IID; blue filled circles, indifference to pain; black open circles, febrile convulsion; black filled circles, Dravet syndrome. *The patient with p.A1632E showed both erythromelalgia and PEPD [21].
