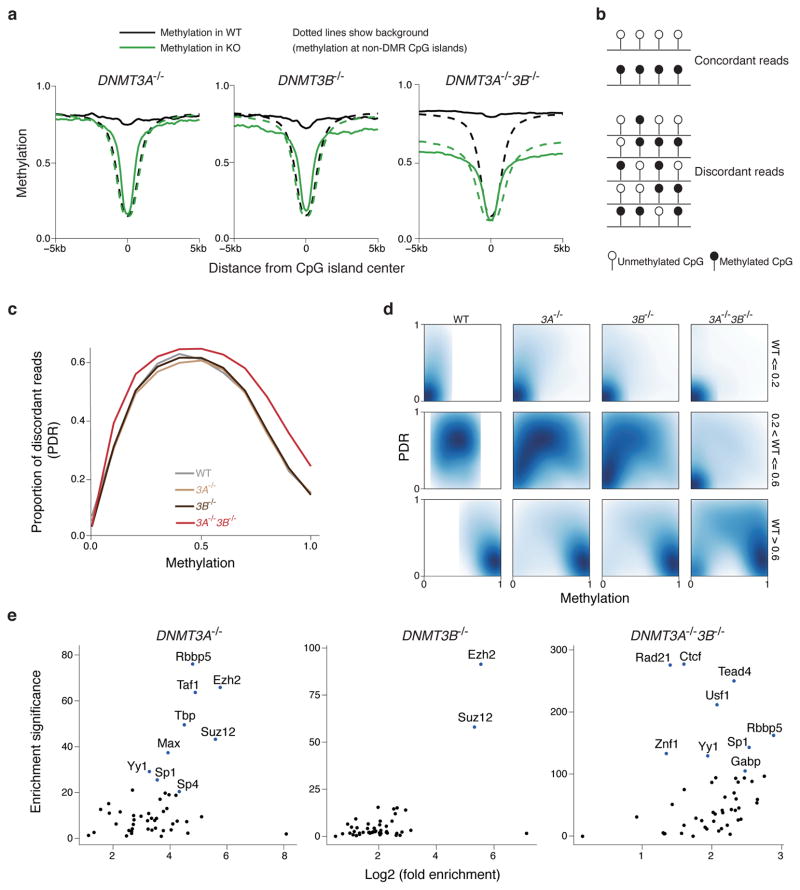
Figure 4. Characterization of targets for DNMT3A and DNMT3B.
(a) Composite plot of methylation around CpG islands in WT and knockouts. The solid lines show the mean methylation of differentially methylated CpG islands, while the dotted lines show the mean methylation of all other CpG islands.
(b) Schematic for the definition of concordant and discordant reads.
(c) Proportion of discordant reads (PDR) in WT, DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells.
(d) Two-dimensional density plots of PDR vs. methylation for CpGs with low (<= 0.2), medium(> 0.2 and <= 0.6) and high(> 0.6) methylation in WT cells.
(e) Enrichment of experimentally determined transcription factor binding sites in H1 ESCs generated by the ENCODE project for DMRs in DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells compared to WT. Enrichment significance was defined as −log10(p-value) of the Fisher’s exact test, with the background being non-repetitive 1kb tiles that had at least 0.4 enrichment in WT.