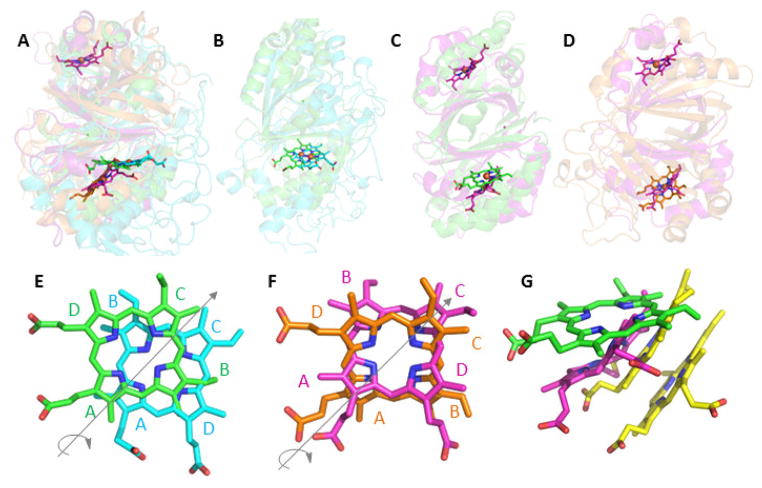
Figure 3. Superimposition of representative CDE protein structures illustrating their diverse heme binding modes.
In each panel, the proteins are shown as cartoons in roughly similar orientations in order to emphasize similarities and differences in the positions of their hemes. The bound hemes are rendered as sticks: Cld (carbon-green, PDB ID 3Q08), DyP (carbon-cyan, 2D3Q), OxdA (carbon-orange, 3W08), IsdG (carbon-magenta, 3LGN), and MhuD (carbon-yellow, 3HX9). A. Superimposition of Cld, DyP, OxdA, and IsdG shows the two distinct planes occupied by the hemes in Cld/DyP and IsdG/OxdA. Note that IsdG is a dimer in which each monomer binds heme, while the other proteins are bidomain monomers where heme binds only in the C-terminal domain. B. Cld/DyP monomer superimposition illustrating the approximate C2 rotation through the heme rings A and C that relates the heme in one of these proteins to the heme in the other. C. Superimposition of the Cld monomer/IsdG dimer. In the lower domain, the side chains on the IsdG heme are oriented similarly to those in DyP, but the heme occupies a different plane. D. Superimposition of the OxdA monomer/IsdG dimer. In the lower domain, the hemes assume roughly the same plane but, once again, are distinguished from one another by a C2 rotation. E–G. Close-up views of the superimposed hemes generated from overlaid CDE protein structures. The heme rings are labeled and the iron atoms have been removed for clarity: E. Cld (green)/DyP (cyan); F. IsdG (magenta)/OxdA (orange); G. Cld (green)/IsdG (magenta)/MhuD (yellow). The side chains of the lower heme in MhuD, an IsdG-family protein from Mycobacterium tuberculosis, are oriented similarly to the heme in IsdG. The upper heme is oriented roughly like the heme in Cld, though in a different plane. Figures generated using PyMol: www.pymol.org.