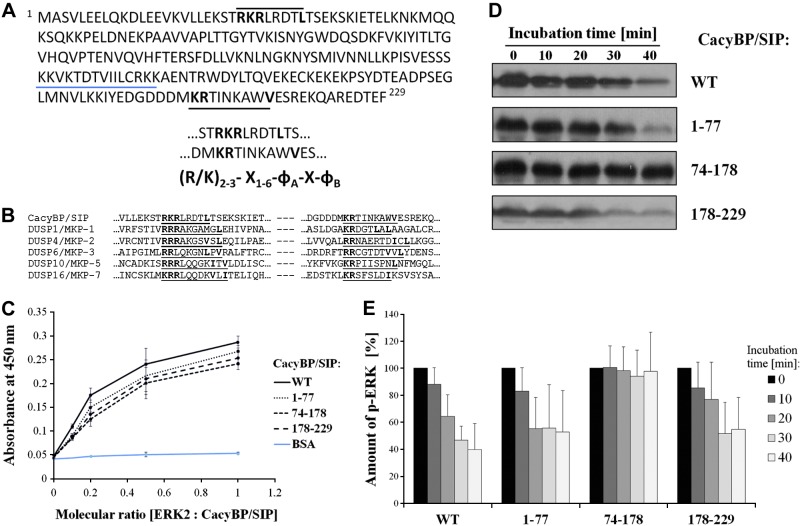
Figure 4.
Analysis of CacyBP/SIP functional organization. A) CacyBP/SIP sequence analysis. Three KIM sequences (underlined), one each in N terminus, CS domain, and C terminus of CacyBP/SIP. X, any amino acid; ɸ, hydrophobic residue: Leu, Ile, Val, ɸB, not obligatory residue. B) Multisequence alignment for CacyBP/SIP and dual-activity phosphatases from MKP family. KIM sequences are underlined. C) ELISA assay for ERK2 binding with full-length CacyBP/SIP wild-type (WT) and domain fragments. D) Phosphatase activity assay for full-length CacyBP/SIP and its fragments, analyzed by Western blot using anti-p-ERK1/2 antibody. E) Densitometric analysis of the activity assay. Statistical analysis was performed for 5 independent experiments.