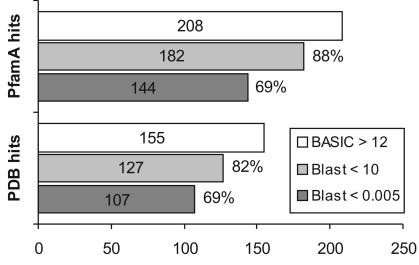
Figure 1.
Statistics of putative annotations obtained with Meta-BASIC for 860 PfamA families of unknown function. The figure shows the number of similarities found with Meta-BASIC between the target families with unknown function (DUF) and other PfamA families or PDB proteins with a conservative Meta-BASIC score cut-off of 12 (BASIC). The bars labelled ‘Blast’ indicate the number of similarities found with Meta-BASIC and supported by Meta-Blast with an E-value below 10 or 0.005, respectively. The Meta-Blast E-values correspond to the lowest E-values reported by PSI-Blast or RPS-Blast.