Figure 6.
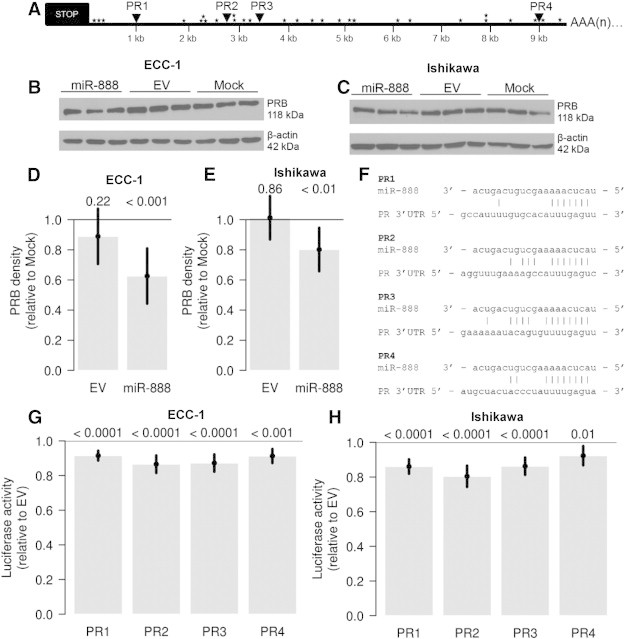
MiR-888 directly targets the PR. (A) PR contains four miR-888 binding sites in its 3′UTR (PR1-PR4, triangles) along with several AREs (stars). Expression of miR-888 was capable of decreasing PR expression at the protein level in both ECC-1 (B) and Ishikawa (C) EC cell lines. Western blots were quantified by densitometry, and PR protein was significantly decreased by miR-888, but not EV transfection, relative to the mock-transfected controls (D: ECC-1, E: Ishikawa). In F, the individual miR-888 binding sites in the PR 3′UTR are aligned with the miR-888 miRNA nucleotide sequence and nucleotide base interactions are denoted with vertical lines. ECC-1 (G) or Ishikawa (H) cells were transfected with luciferase reporters containing each PR 3′UTR miR-888 binding site (PR1-PR4) downstream of Renilla luciferase in combination with miR-888 or the EV control. MiR-888 expression resulted in a significant decrease in luciferase activity compared to the EV control for all four miR-888 binding sites (G: ECC-1, H: Ishikawa). For D, E, G, and H, statistical significance was determined using ANOVA models and error bars represent 95% confidence intervals. P values are listed above each bar graph.
