Figure 4.
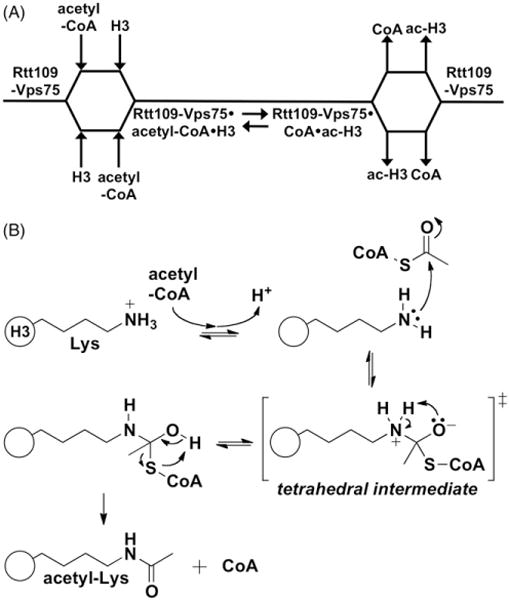
Current model for enzymatic mechanism of yeast Rtt109. (A) Cleland diagram for Rtt109-Vps75-catalyzed histone acetylation depicting the proposed order of substrate binding and product release. H3 is depicted as the substrate, but it can most likely include other substrates. Protein substrate and acetyl-CoA appear to bind and depart Rtt109-Vps75 in a non-obligate order. (B) Proposed catalytic model of Rtt109-catalyzed histone acetylation. The lysine substrate (pKa ~10) becomes deprotonated during the course of binding. In the absence of a general base catalyst, the lysine side chain is likely positioned at an optimal distance and orientation with respect to bound acetyl-CoA to undergo a nucleophilic attack at the acetyl carbonyl carbon. A tetrahedral intermediate would then be formed, followed by product release. The protein and/or solvent component(s) that likely stabilize the transition states are unknown.
