Figure 1.
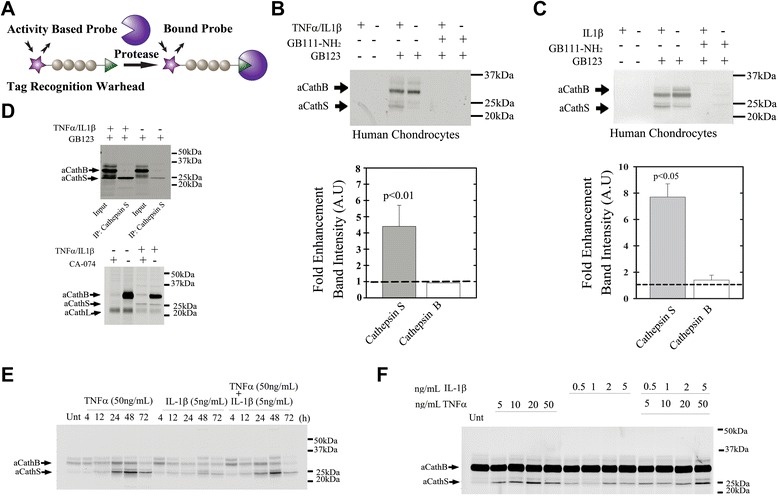
Activity-based probe (ABP) targets active cathepsins in TNF α/IL1 β-stimulated chondrocytes. (A) ABP binding mechanism. Illustration shows the general structure of an APB with a triangular reactive moiety. Covalent modification of the target protease results in production of a fluorescently labeled enzyme. (B) Protein extracts from TNFα/IL1β-treated and untreated chondrocytes labeled with GB123 with or without pretreatment of cathepsin inhibitor GB111-NH2, as indicated (n = 4) with quantification of band intensity (Image J software) below. (C) Protein extracts from IL1β-treated and untreated chondrocytes labeled with GB123 with or without pretreatment of cathepsin inhibitor GB111-NH2, (n = 4) with quantification below. (D) Protein extracts from TNFα/IL1β-treated and untreated chondrocytes were labeled with GB123 and immunoprecipitated with cathepsin S antibody (n = 4). Below, a gel showing protein extracts incubated with or without CA-074 (selective cathepsin B inhibitor) and labeled with GB123. The long chain of active Cathepsin L was detected between 20 and 25 kDa. (E) Time-course experiments (50 ng/mL TNFα and 5 ng/mL IL1β) with GB123 labeling; experiment duration indicated above the gel (n = 4). (F) Dose response experiments (24-h incubation), with GB123 labeling, performed for cytokine concentrations (ng/mL);(n = 4). Gels were scanned (Typhoon scanner Excitation/Emission 630/670 nm) to detect fluorescent-labeled active cathepsin bands. aCathB, active cathepsin B (32 kDa); aCathS, active cathepsin S (25 kDa); IP, immunoprecipitation, Unt, untreated. Band intensity plots (image J software) show SD around average data point. Statistical analysis was by non-parametric Mann-Whitney analysis, considering P <0.05 statistically significant.
