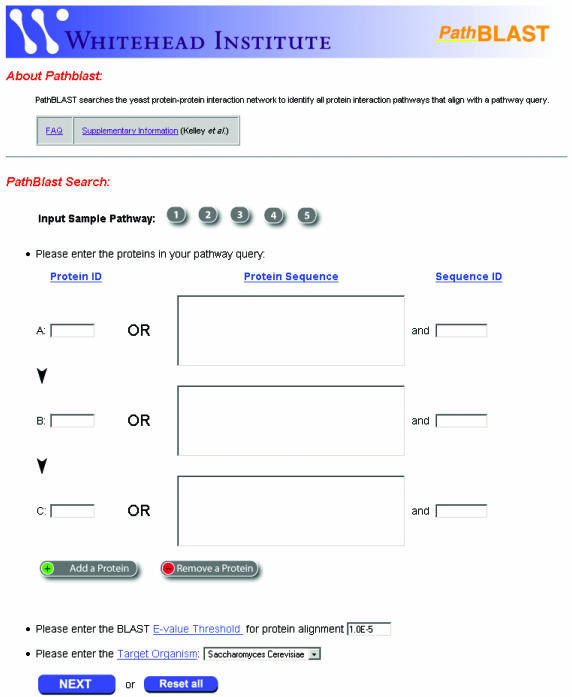
Figure 2.
PathBLAST front page. To define the pathway query, users enter a series of protein IDs (DIP number, common name, or systematic ORF designation), or a series of FASTA-format protein sequences, each with a corresponding sequence identifier. The length of the query pathway can be varied between two and five proteins by using the ‘Add a Protein’ and ‘Remove a Protein’ buttons. Users must specify the BLAST E-value threshold for protein sequence similarity (used to determine which protein pairs should be considered as putative orthologs) as well as the target network for comparison. As part of a brief tutorial, users can evaluate the approach on several example pathways.