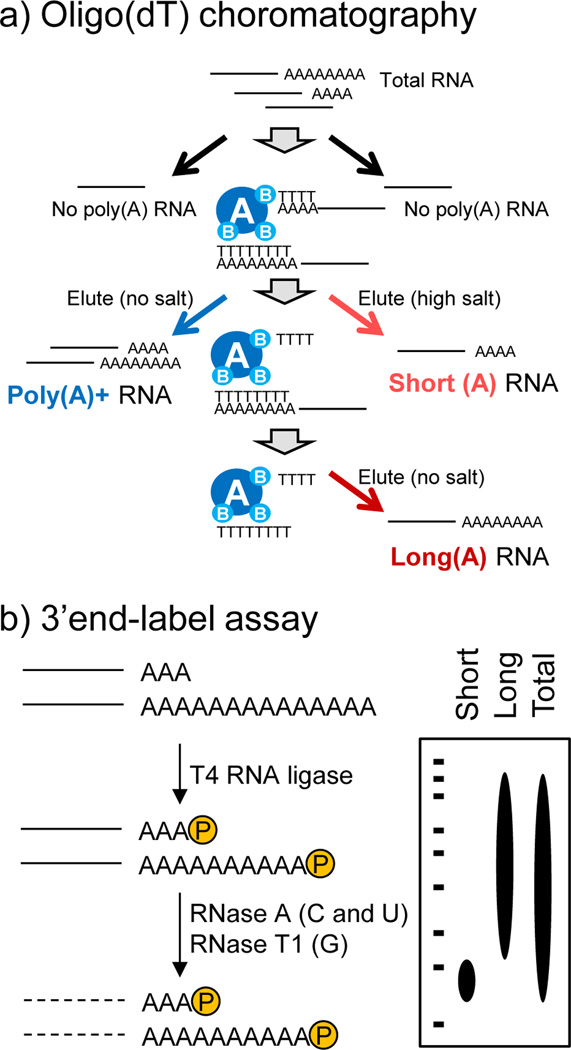
Figure 1. Method for fractionation of mRNAs with different poly(A) tail lengths (Kojima et al., 2012).
a) Oligo(dT) chromatography was used to separate total RNA into fractions with either short or long poly(A) tails by varying salt concentrations in the elution. Total PolyA+ (non-fractionated) RNAs can also be isolated as a reference. b) Validation of fractionation by a 3’-end label assay. T4 RNA ligase catalyzes the ligation of the 5' phosphate terminus of a nucleic acid donor to the 3' OH terminus of a nucleic acid acceptor, thus introduces radioisotope label at the 3’-end of poly(A) tails. 3’-end labeled mRNAs are then digested by both RNase A that recognizes cytidine and uridine residues and RNase T1 that recognizes guanidine residues, resulting in degradation of the RNA body but not poly(A) stretches. The size of these poly(A) stretches can be visualized by autoradiography after PAGE (polyacrylamide gel electrophoresis) analysis.