Summary
The spatiotemporal balance between stem cell maintenance, proliferation, and differentiation determines the rate of root growth and is controlled by hormones, including auxin and brassinosteroid (BR). However, the spatial actions of BR and its interactions with auxin remain unclear in roots. Here, we show that oppositely patterned and antagonistic actions of BR and auxin maintain the stem cell balance and optimal root growth. We discover a pattern of low levels of nuclear-localized BR-activated transcription factor BZR1 in the stem cell niche and high BZR1 levels in the transition-elongation zone. This BZR1 pattern requires local BR catabolism and auxin synthesis, as well as BR signaling. Cell-type-specific expression of a constitutively active form of BZR1 confirms that the high and low levels of BZR1 are required for the normal cell behaviors in the elongation zone and quiescent center (QC), respectively. Comparison between BR-responsive, BZR1-targeted, auxin-responsive, and developmental zone-specific transcriptomes indicates that BZR1 mostly activates its target genes expressed in the transition-elongation zone, but represses genes in the QC and surrounding stem cells, and that BR and auxin have overall opposite effects on gene expression. Genetic and physiological interactions support that a balance between the antagonistic actions of BR and auxin is required for optimal root growth. These results demonstrate that the level and output specificity of BR signaling are spatially patterned and that, in contrast to their synergism in shoots, BR and auxin interact antagonistically in roots to control the spatiotemporal balance of stem cell dynamics required for optimal root growth.
Graphical abstract
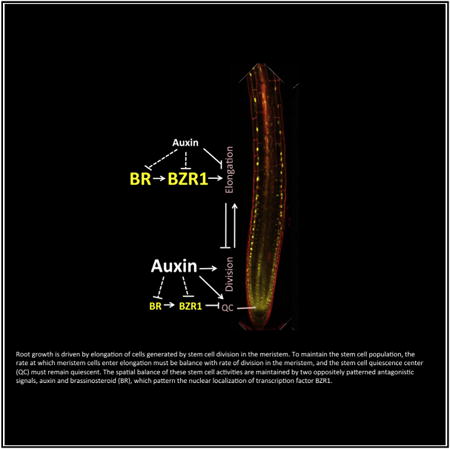
Introduction
The spatial balance between stem cell maintenance, proliferation, and differentiation determines the pattern and rate of growth in multicellular organisms. The continuously growing root tip contains a population of cells organized in a radially symmetrical pattern with a longitudinal gradient of quiescence, proliferation, and elongation. At the apical side of the root meristem, stem cells within the quiescent center (QC) rarely divide but specify the identity of the surrounding initial cells, which divide to form the meristem zone. At the basal side of the meristem, cells exit the meristem zone through a transition zone and enter the elongation zone, where cells stop division but rapidly increase their length and volume [1]. Both cell division in the meristem zone and cell elongation in the elongation zone contribute to root growth [2], and their balance is crucial for sustained and optimal root growth and is regulated by several hormones [3].
One of the most important hormones for root development is auxin. Auxin is known to induce root initiation in explants, promote lateral root development, and inhibit root elongation [3]. Both localized synthesis and active polar transport establish an auxin gradient with maxima at the QC [4, 5]. High levels of auxin maintain stem cell activities in the stem cell niche (QC and surrounding initial cells) to regulate meristem size, partly by activating the transcription factors PLETHORA [6, 7]. The low auxin level in the elongation zone allows cell elongation, and auxin inhibition of cell elongation mediates gravitropic root bending [3, 8, 9]. Auxin activates the auxin response transcription factors (ARFs) by triggering degradation of the Aux/IAA family transcriptional repressor proteins [10]. Recently, the auxin-responsive transcriptome has been shown to vary along the developmental zones of root tips [11]. However, the mechanisms by which auxin induces developmental zone-specific responses remain unknown.
Another important regulator of root growth is brassinosteroid (BR). BR-deficient or BR-insensitive mutants exhibit a wide range of growth defects, including dwarf shoot and short root [12]. Although BR stimulates cell elongation in shoot organs, its effect on root growth is dose dependent [13, 14]. Recent studies showed that BR at different concentrations can have opposite effects on root meristem size by promoting both cell-cycle progression and cell differentiation [15]. BR also modulates cell division in the QC [15–18], and BR signaling in the epidermis is sufficient for promoting root growth [17]. However, it remains unclear whether endogenous BR is spatially patterned to differentially regulate stem cell activities in the root tip.
BRs bind to the extracellular domain of the receptor kinase BRI1 to trigger a cascade of downstream signal transduction events, which include inactivation of the GSK3-like kinase BIN2, dephosphorylation of the BRASSINAZOLE RESISTANT 1 (BZR1) family transcription factors by PP2A phosphatase, accumulation of unphosphorylated BZR1 in the nucleus, and BZR1 regulation of thousands of BR target genes [19]. Many studies have demonstrated the synergistic effects of BR and auxin in several developmental processes, such as hypocotyl elongation, vascular differentiation, and lateral root initiation, as well as in gene expression [12, 20–24]. However, there has been conflicting genetic evidence supporting synergistic or antagonistic BR-auxin interaction in root growth [25–28]. Although interactions between signaling components of the BR and auxin pathways have been observed [29, 30], the functions of these interactions in root tip growth remain unknown.
Here, through microscopic, transcriptomic, and genetic analyses, we demonstrate that BZR1 is activated by endogenous BR in a graded pattern along developmental zones, which is overall opposite to the auxin gradient in root tips, and that BR acts antagonistically with auxin on BZR1 nuclear localization, transcriptomic response, and cell elongation in developmental zone-specific manners. Our study demonstrates that the stem cell dynamics are controlled by oppositely patterned antagonistic actions of BR and auxin, mediated at least in part by BZR1, in Arabidopsis root tips.
Results
The Positive and Negative Effects of BR on Root Growth Are Separated Spatiotemporally and Are Concentration Dependent
Previous studies demonstrated that BR promotes root growth at low concentrations but inhibits root growth at high concentrations [13–15] (Figure 1A). The promoting effect of BR, however, is subtle and sometimes undetectable in wild-type plants but is obvious in the BR-deficient mutant dwf4, which shows a severe short-root phenotype that can be rescued by 50 pM brassinolide (BL; the most active form of BR) (Figure 1A), suggesting that under our experimental conditions, wild-type plants maintain near-optimal endogenous BR levels for root growth.
Figure 1. Brassinosteroid Regulates Dynamics of Root Growth in Arabidopsis.
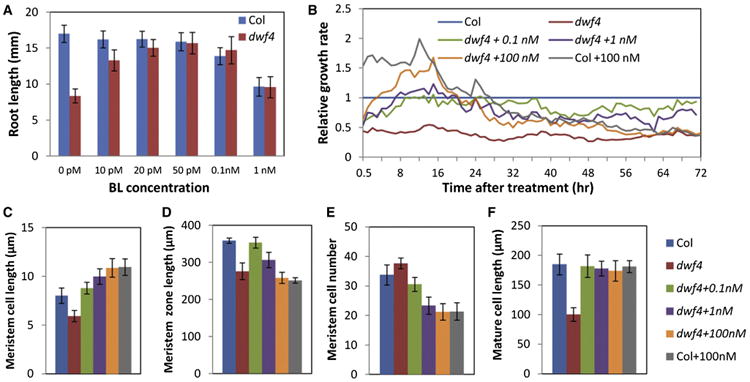
(A) Root growth measurement of 6-day-old wild-type (Col) and dwf4 seedlings grown on media with various concentrations of BL (n > 20, mean ± SD).
(B) Average root growth rates of Col and dwf4 seedlings after being transferred to media containing BL or mock were measured over time by live imaging and normalized to Col (n > 10).
(C–F) Measurement of cortical meristem cell length (average length of cell numbers 1–15 from cortex-endodermis initials) (C), meristem zone length (D), meristem cell number (E), and mature cell length (F) after treatment with BL or mock for 24 hr (n > 12, mean ± SD).
See also Figure S1.
To understand how BR modulates root growth dynamics, dwf4 seedlings were treated with a range of concentrations of BL, and the root growth rates were measured using live imaging over 72 hr. Untreated dwf4 roots grew on average at about 40% the speed of wild-type roots (Figure 1B). Immediately after treatment with BL, root growth was accelerated. However, after about 12 hr, the growth rates of roots treated with high concentrations of BL (e.g., 100 nM) started to decline gradually and became lower than untreated wild-type control roots by about 24 hr (Figures 1B and S1). Microscopic analysis of the root tips after 24 hr of treatment revealed that low BL concentration (0.1 nM) rescued the size and cell organization of meristem and the mature cell length in dwf4 roots (Figures 1C–1F). In contrast, high concentrations of BL (1 nM and 100 nM) promoted cell elongation in the meristem (Figure 1C) and reduced meristem size (length and cell number) (Figures 1D and 1E) but did not further increase mature cell size (Figure 1F). Such kinetics of BR response confirm that an increase of BR concentration promotes cell elongation and exit from the meristem [15], leading to reduced meristem size and growth rate after an initial growth increase due to cell elongation.
BR Signaling Is Patterned in Root Tip
BR signaling increases BZR1 nuclear localization through PP2A-mediated dephosphorylation [31–33]. Therefore, the level of nuclear BZR1 provides a readout of BR signaling. The BZR1-YFP fusion protein expressed from either the BZR1 promoter or the constitutive 35S promoter accumulated at a low level in the nuclei of stem cell region but at a higher level in the nuclei of epidermal cells in the transition and elongation zone, in contrast to the histone-RFP protein expressed in the same plant (Figures 2A and 2B; Figure S2; Movie S1). The non-fusion GFP protein expressed from the BZR1 or 35S promoter showed a more uniform distribution in the root tip (Figures S3A and S3B), whereas BES1/BZR2-GFP showed a similar pattern as BZR1-YFP with high nuclear intensity in the epidermal cells of transition-elongation zone (Figures S3C and S3D). Quantification of the nuclear and cytoplasmic BZR1-YFP signals revealed a gradual decrease of cytoplasmic BZR1 from root tip to the elongation zone and a rapid increase of nuclear BZR1 level as cells entered the transition-elongation zone (cell length to width [l/w] ≥ 1.0), correlating with rapid cell elongation (Figure 2C). The nuclear-to-cytoplasmic (N/C) ratio of BZR1-YFP showed a gradient increase along the root developmental axis (Figures 2D and S3E–S3I). In addition, BZR1 accumulated to a high level in the phloem (Figure S3J), which correlates with the role of BR in vascular differentiation and BZR1 regulation by the TDIF peptide signal [34, 35].
Figure 2. BZR1 Activity Is Regulated in a Spatiotemporal Manner by BR Biosynthesis and Catabolism in Roots.
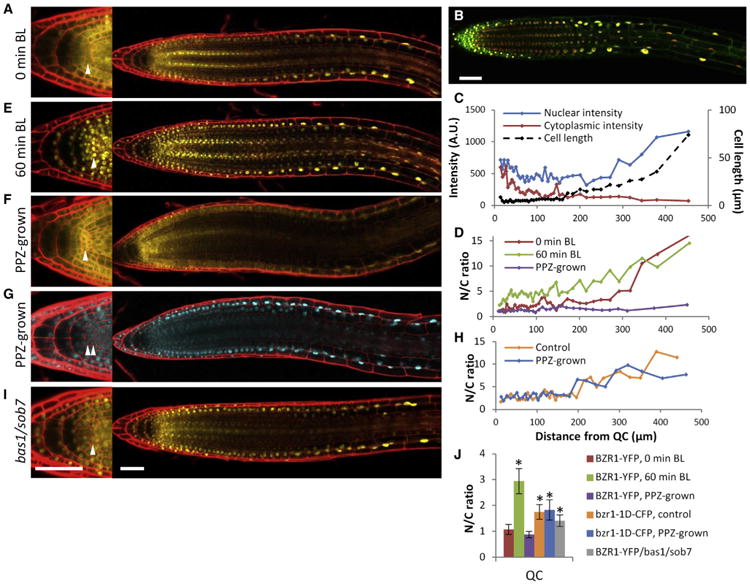
(A) Confocal images of BZR1-YFP localization in a root tip of the pBZR1:BZR1-YFP plant.
(B) Overlay of confocal images of BZR1-YFP (green) and H2B-RFP (red) expressed from the 35S promoter in the root of a transgenic plant grown on regular medium.
(C) YFP intensities and cell lengths of each epidermal cell along the root axis before BL treatment.
(D–F) N/C ratios (D) of the roots before (A) and after (E) BL treatment (100 nM BL for 60 min) or grown on 2 μM PPZ medium (F).
(G and H) bzr1-1D-CFP localization (G) and N/C ratios (H) in root tips of pBZR1:bzr1-1D-CFP × 35S:H2B-RFP seedlings when grown on 2 μM PPZ medium (G) or regular medium (Figure S3M).
(I) pBZR1:BZR1-YFP/bas1-2;sob7-1 grown on regular medium.
(J) N/C ratios of BZR1-YFP or bzr1-1D-CFP in QC cells (n > 5, mean ± SD). *p < 0.001, as determined by a Student's t test.
Magnifications of the stem cell niche and columella are shown on the left (A, E–G, and I). Arrowheads indicate QC cell layers. Scale bars represent 50 μm. Roots were stained with propidium iodide (red) in panels (A), (E)–(G), and (I). See also Figures S2 and S3 and Movie S1.
Consistent with nuclear BZR1-YFP reflecting the pattern of BR signaling, BR treatment (100 nM BL, 60 min) promoted BZR1 nuclear localization (increased the N/C ratio) in all cells except the cells in the elongation zone that have maximum N/C ratio without exogenous BR (Figures 2D, 2E, and S3E–S3I). By contrast, treatment with a BR biosynthesis inhibitor (propiconazole, PPZ), as well as the dwf4 and bri1-116 mutations, reduced nuclear localization of BZR1 in all cell types, especially the epidermis of transition-elongation zone (Figures 2D, 2F, S3K, and S3L), causing a BZR1-YFP pattern similar to GFP alone (Figure S3B). These results suggest that the gradient pattern of BZR1 in the root tip is due, at least partly, to patterned distribution of endogenous BR.
The gain-of-function bzr1-1D mutation increases BZR1's affinity to PP2A phosphatases, resulting in an activated and hypophosphorylated form of BZR1 [33]. In the root tip, the bzr1-1D-CFP fusion protein was detectable in the nucleus of all cell types and caused extra QC division even when BR biosynthesis was blocked (Figures 2G, 2H, 2J, S3M, and S3N). Consistently, the bzr1-1D-CFP and bzr1-1D mutation also partly rescued the short-root phenotypes of dwf4 and bri1-116, respectively (Figure S3O), suggesting that BR regulates root growth mostly through BZR1-mediated transcriptional responses.
The Gradient of Nuclear BZR1 Does Not Require Localized BR Synthesis but Involves Localized BR Catabolism
When dwf4 plants expressing BZR1-YFP protein were grown on media containing 50 pM BL, a normal pattern of BZR1-YFP was observed (Figure S3P), suggesting that the BZR1 pattern can be established from exogenous BL, either by BL catabolism or transport or differential BR sensitivity, without localized BR synthesis. In the loss-of-function mutant of BR catabolic enzymes bas1-2;sob7-1 (BAS1/CYP72B1 and SOB7/CYP72C1) [36], an increased nuclear BZR1-YFP signal was observed in the stem cells and columella cells (Figure 2I), indicating that BR catabolism is required for maintaining a low BR level and cytoplasmic BZR1 localization in the stem cell region of the root tip. Promoter-GFP reporter lines showed that both BAS1 and SOB7 are expressed in the root cap (Figures S3Q and S3R), which might reduce availability of bioactive BR in the adjacent columella and stem cell niche. There is also a low level of SOB7 expression in the epidermis of the elongation zone, which may mediate feedback regulation of BR homeostasis to maintain optimal cell elongation dynamics.
BZR1 Has Cell-Type-Specific Functions in Root Tip
To examine the role of spatiotemporal BZR1 activity in root development, we manipulated BZR1 activity in specific cell types by expressing bzr1-1D-YFP under cell-type-specific promoters in the bri1-116 background. The pWER:bzr1-1D-YFP construct, which expressed activated BZR1 in the epidermis, rescued the sizes of meristem zone, elongation zone, and mature cells, and partially rescued the short-root phenotype of bri1-116 (Figures 3A–3F). However, expression of activated BZR1 in the QC by pWOX5:bzr1-1D-YFP and in the endodermis and QC by pSCR:bzr1-1D-YFP had no effect on meristem size, cell elongation, or root length (Figures 3E–3H) but increased division of QC cells (Figures 3G–3L). The findings suggest that BZR1 acts in the epidermis to promote root meristem growth and root cell elongation but can promote QC division in a cell-autonomous manner.
Figure 3. Cell-Type-Specific BZR1 Activities Modulate Normal Root Development.
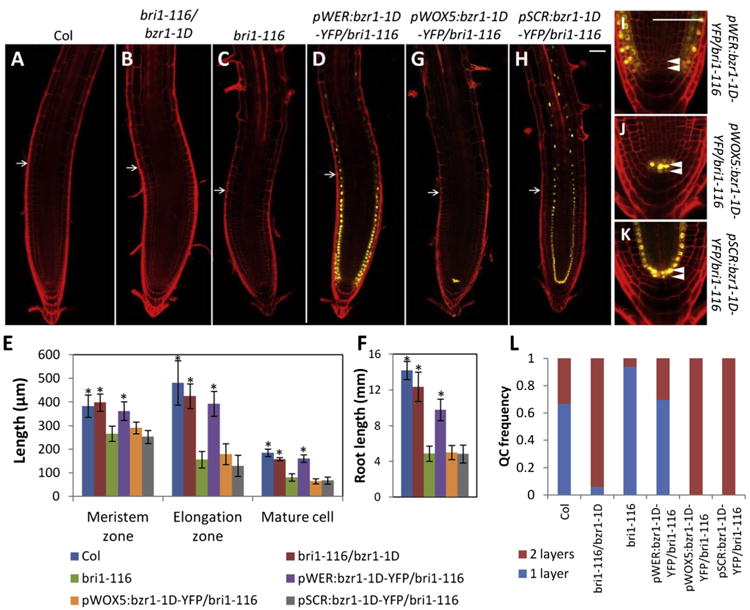
(A–D and G–K) Confocal microscopic analysis of root tips of Col, bri1-116/bzr1-1D, bri1-116, and transgenic lines expressing bzr1-1D-YFP under the indicated cell-type-specific promoters in bri1-116 background.
(E, F, and L) Quantification of meristem zone length, elongation zone length, mature cell length (E), root length (F), and QC layers (L). (n > 20, mean ± SD).
*p < 0.001, as determined by a Student's t test. Arrows indicate transition zone. Arrowheads indicate QC cell layer (I–K). Scale bars represent 50 μm.
The Pattern of Nuclear BZR1 Is Modulated by Auxin
An auxin gradient, with a maximum in the root tip established by both polar auxin transport and localized synthesis by tryptophan aminotransferase TAA1, controls developmental zonation of stem cell maintenance and cell elongation in roots [37, 38]. To test whether auxin is involved in establishing BR's spatial pattern, we analyzed BZR1-YFP localization pattern in root tips treated with auxin (indole-3-acetic acid, IAA) or an auxin biosynthesis inhibitor, L-kynurenine, which inhibits TAA1 [39]. Treatment with auxin caused cytoplasmic accumulation of BZR1-YFP in the epidermal cells of the transition-elongation zone after 5 hr and decreased nuclear accumulation after 24 hr but had no obvious effect on BZR1 accumulation in the meristem (Figures 4A–4D, S4A, and S4B). Short-term auxin treatments (<4–5 hr) or gravity-induced auxin redistribution, however, did not significantly alter the BZR1 localization pattern (Figure S4C). In turn, L-kynurenine inhibition of local auxin biosynthesis in the QC area [38, 39] led to increased nuclear accumulation of BZR1-YFP in the QC and columella stem cells (Figures 4E, 4F, and S4D– S4G). The results suggest that auxin inhibits BZR1 nuclear accumulation and the auxin gradient contributes to the normal BZR1 patterning in the root tip.
Figure 4. The Pattern of Nuclear BZR1 Is Modulated by Auxin.
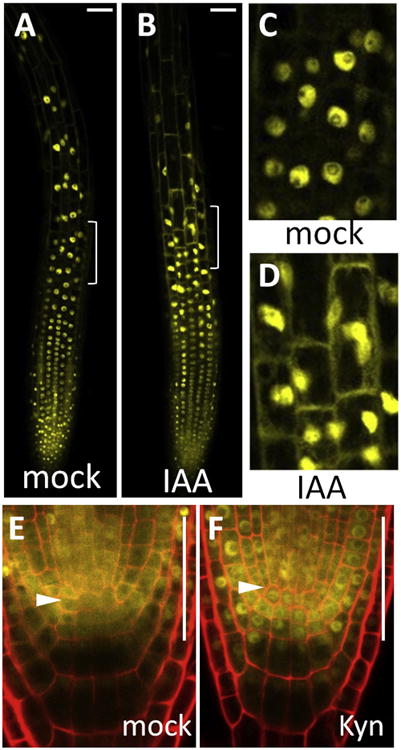
(A–D) BZR1-YFP localization pattern in the epidermis after 5 hr treatment with mock (A and C) or 5 μM IAA (B and D).
(C and D) Zoomed-in views of the transition zone in (A) and (B) as indicated by white brackets.
(E and F) BZR1-YFP localization pattern in the QC after 24 hr treatment with mock (E) or 25 μM auxin biosynthesis inhibitior (Kyn) (F). Arrowheads indicate QC cell layer.
Scale bars represent 50 μm. See also Figure S4.
BR and Auxin Antagonistically Regulate Gene Expression in Root
To understand the function of BZR1 and BR-auxin interaction in root development, we performed RNA sequencing (RNA-seq) experiments to identify BR- and auxin-responsive genes in root tips. Combining genes affected significantly (p < 0.01, log2 fold change > 0.5 [fold change > 1.4]) by BR treatment (Col + BL versus Col and dwf4 + BL versus dwf4; 100 nM BL) and BR mutations (Col versus dwf4, Col versus bri1-116, and bri1-116,bzr1-1D versus bri1-116), we identified 2,966 BR-induced genes and 2,344 BR-repressed genes (Table S1; Figure S5A). About 65% of genes affected by the bri1-116 mutation were affected by bzr1-1D, and 99% of these were affected in opposite ways by bzr1-1D (Figure S5B), consistent with bzr1-1D suppressing phenotypes of bri1-116 (Figure S3O). About 34% of the BR-regulated genes were direct targets of BZR1 identified in previous BZR1 chromatin immunoprecipitation (ChIP)-chip or chromatin immunoprecipitation sequencing (ChIP-seq) experiments [24, 40] (Table S2). We identified 2,806 and 3,099 genes induced and repressed by auxin treatment (5 μM IAA), respectively (p < 0.01, log2 fold change > 0.5; Table S1). These include about 52% of the BR-regulated genes (Figure 5A), indicating a high degree of coregulation. Surprisingly, in contrast to previous observation of similar effects of BR and auxin on overlapping transcriptome in whole seedlings [22, 23], more than 70% of the coregulated genes responded to BR and auxin in opposite directions (Figure 5B; Table S2). Co-treatment of wild-type seedlings simultaneously with BR and auxin caused an overall transcriptomic change similar to auxin treatment alone, but with reduced fold change for the genes oppositely affected by individual BR and auxin treatments and enhanced response for the genes similarly regulated by each hormone (Figures 5B and S5C). qRT-PCR analysis of five BZR1 target genes confirmed their variable coregulation by BR and auxin (Figure 5C).
Figure 5. BR and Auxin Regulate Overlapping Transcriptomes.
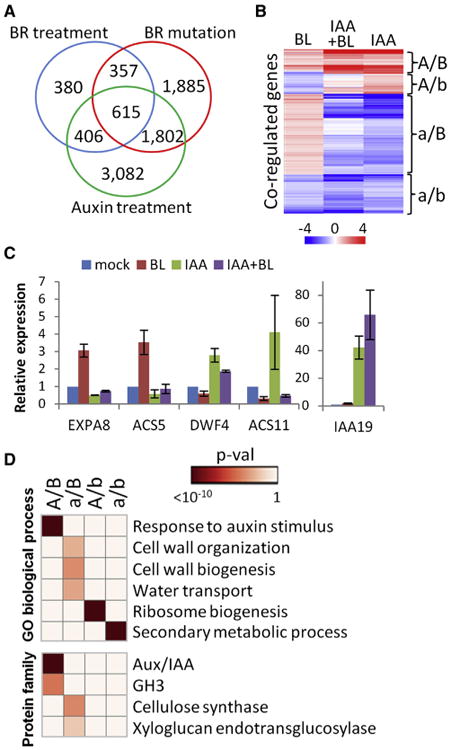
(A) Venn diagram of genes affected by BR treatment, BR mutation, or auxin treatment.
(B) Hierarchical cluster analysis of log2 fold change values of BR-auxin coregulated genes after 4 hr treatment of Col with IAA, BL, or IAA + BL.
(C) qRT-PCR analysis of expression of BR-auxin coregulated BZR1 target genes (n = 3, mean ± SD).
(D) Enriched GO and protein family categories of different BR-auxin coregulated genes.
A/B = auxin induced, BR induced; a/B = auxin repressed, BR induced; A/b = auxin induced, BR repressed; a/b = auxin repressed, BR repressed. Heatmap shows p values of the enriched terms. See also Figure S5 and Table S1.
Enrichment analysis of gene ontology (GO) and InterPro functional categories indicates that both BR and auxin induce genes involved in response to auxin stimulus, including many Aux/IAA and GH3 family genes (Figure 5D). The BR-induced but auxin-repressed genes tend to be involved in cell-wall organization and biogenesis and water transport, suggesting that BR and auxin have opposite effects on cell elongation in root. BR-repressed but auxin-induced genes tend to be involved in ribosome biogenesis, and genes repressed by both BR and auxin tend to be involved in secondary metabolic processes (Figure 5D). These results suggest that BR and auxin are largely antagonistic, but their relationship varies for regulating different cellular functions in root.
BR and Auxin Are Major Regulators of Developmental Zone-Specific Gene Expression in Primary Root
Previous studies have provided a high-resolution RootMap expression profile of several cell types and longitudinal sections of a single root [41]. Comparison of BR-responsive gene expression data with the RootMap gene expression data shows a distinct pattern of BR activation versus repression of genes specifically enriched in different developmental zones (Figure 6A; Table S2). Our auxin-responsive gene sets also showed developmental zone-specific responses (Figure 6A), consistent with that reported recently [11]. There is overall a strong inverse correlation between gene responses to BR and auxin along the developmental zones (Figures 6A and 6B). In particular, 41% of the QC-enriched genes [11] are repressed and only 10% of them are induced by BR (Figure 6C), and 37% of the BR-repressed genes are induced and only 15% are repressed by auxin. Similarly, genes enriched in the meristem zone are more likely repressed (13%) than induced (3%) by BR, and about 43% of BR-repressed genes in the meristem zone are induced by auxin, and only 14% of them are also repressed by auxin. In contrast, genes expressed in the transition-elongation zone are more likely induced (36%) than repressed (7%) by BR, and 65% of the BR-induced genes in the transition-elongation zone are repressed, and only 6% of them are induced by auxin (Figure 6C). Genes expressed in late elongation and differentiation zones showed less bias toward activation or repression by the hormones but still showed the overall trend of being regulated by BR and auxin in opposite ways (Figures 6B and 6C; Table S2).
Figure 6. BR and Auxin Regulate Gene Expression in a Developmental Zone-Specific Manner.
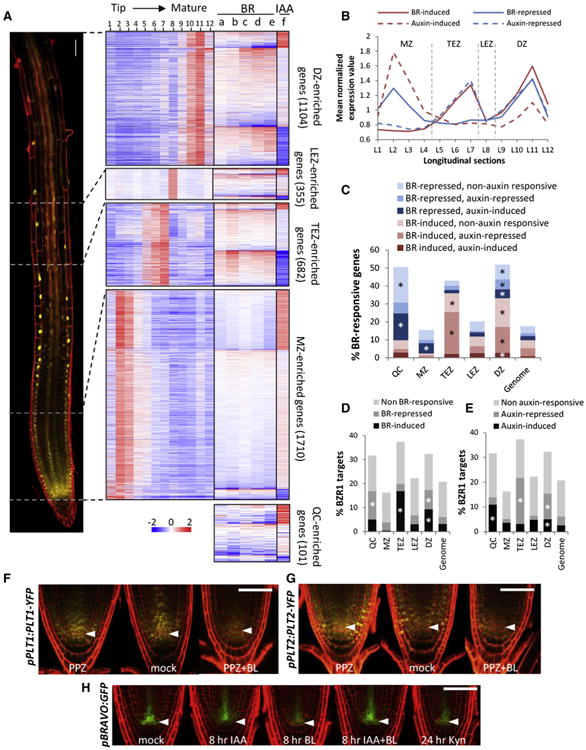
(A–E) BR and auxin responses of genes specifically enriched in the QC, meristem zone (MZ), transition-elongation zone (TEZ), late elongation zone (LEZ), and differentiation zone (DZ) based on the RootMap gene expression dataset [41].
(A) Heatmaps show log2 fold change values of genes specifically enriched in QC and different developmental zones from indicated BR and auxin RNA-seq experiments: Col + BL versus Col (a), dwf4 + BL versus dwf4 (b), Col versus dwf4 (c), Col versus bri1-116 (d), bri1-116/bzr1-1D versus bri1-116 (e), and Col + IAA versus Col (f). Mean-normalized expression values (log2 transformed) of zone-enriched genes from 12 sections along the root longitudinal axis [41] are shown in the same row order with their corresponding BR-auxin expression values. pBZR1:BZR1-YFP root tip divided into four developmental zones is shown on the left.
(B) Average mean-normalized expression values of developmental zone-enriched, BR- and auxin-responsive genes in 12 longitudinal sections (L1–L12).
(C–E) Percentage distribution of BR-responsive genes and their auxin response (C) and of BZR1 target genes and their BR response (D) or auxin response (E) in QC and different developmental zones, compared to the whole genome. Asterisk (*) indicates significant overrepresentation (hypergeometric p value < 0.0001).
(F–H) Expression of pPLT1:PLT1-YFP (F) and pPLT2:PLT2-YFP (G) in roots grown on mock or 2 μM PPZ medium for 6 days and transferred to 2 μM PPZ medium with or without 100 nM BL for 24 hr.
(H) Expression of pBRAVO:GFP after treatments of 5 μM IAA, 100 nM BL, or both for 8 hr, or 25 μM Kynurenine for 24 hr.
Arrowheads indicate QC cell layer. Scale bars represent 50 μm. See also Table S2.
The dominant groups of BR/auxin-coregulated genes represent distinct functions in different developmental zones. For example, 46 out of the 97 BR-repressed auxin-induced meristem genes are related to ribosome biogenesis (Table S3). Loss of function of some of these genes has been shown to cause root growth defects [42, 43]. By contrast, BR-induced and auxin-repressed genes of the transition-elongation zones are highly enriched with functions in cell-wall biogenesis (p < 0.001).
Similar to BR-responsive genes, the BZR1 target genes are also overrepresented among the genes expressed in the QC (32%), transition-elongation zone (36%), and differentiation zone (32%), but underrepresented among genes expressed in the meristem zone (16%) (Figure 6D). The BZR1 target genes expressed in the QC tend to be repressed by BR and induced by auxin, whereas those expressed in the transition-elongation zone tend to be induced by BR but repressed by auxin (Figures 6D and 6E). Among the BR-repressed and auxin-induced BZR1 target genes are many major regulators of root development, including PLT1, PLT2, PLT4/BBM, and BRAVO (Table S2), which were reported recently as key factors required for stem cell maintenance [16, 44]. Consistent with the RNA-seq data, the levels of PLT1-YFP and PLT2-YFP expressed from each native promoter were reduced by BR treatment (Figures 6F and 6G). The RNA level of BRAVO was increased by auxin treatment (Table S1; [11]), but the pBRAVO:GFP reporter did not respond to auxin treatment (Figure 6H; [16]). However, auxin increased pBRAVO:GFP expression in the presence of BR, and inhibiting auxin biosynthesis reduced the pBRAVO:GFP level (Figure 6H). These results suggest that the endogenous auxin maximum is required and sufficient for BRAVO expression under normal conditions but becomes insufficient when the BR level is elevated in the QC and surrounding area.
BR and Auxin Antagonistically Regulate Cell Elongation and Root Growth
The complementary spatial patterns of activity and opposite effects on gene expression suggest that BR and auxin function oppositely in regulating root growth. Indeed, roots of the BR-deficient mutant dwf4 were hypersensitive to auxin, and BL reduced the auxin sensitivity of dwf4 in a dose-dependent manner (Figure 7A). Conversely, auxin reduced BL sensitivity of dwf4 roots. Treatment with 60 nM 2,4-D (a synthetic auxin) shifted the optimal BL concentration for root growth from 0.05 to 0.5 nM (Figure 7B), indicating that the balance between the two antagonistic hormones is critical for optimal root growth. Furthermore, the dominant bzr1-1D mutation was able to suppress the auxin hypersensitivity of dwf4 and the BR signaling mutants bri1-116 and bin2-1 (Figures 7C and S6), consistent with BZR1 being involved in the BR-auxin interaction in root growth.
Figure 7. BR and Auxin Antagonistically Regulate Growth Dynamics in Root Tip.
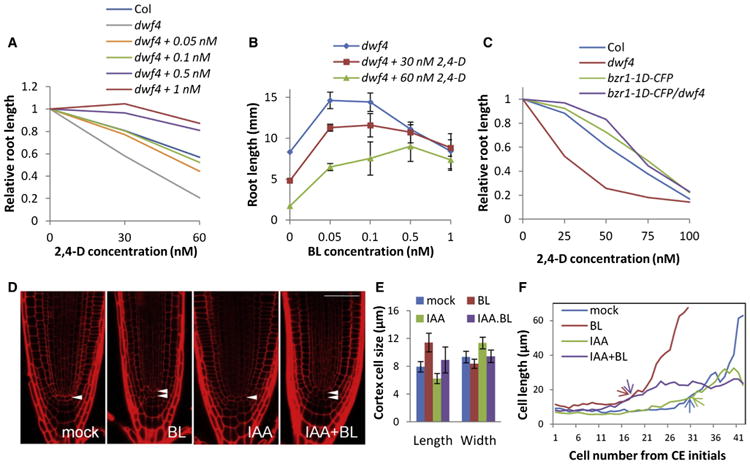
(A–C) Measurement of root length of 6-day-old seedlings of indicated genotype grown on vertical media supplemented with various concentrations of 2,4-D and/or BL. Relative root length is shown in (A) (n > 15) and (C) (n > 15). The same data in (A) were used in (B) (mean ± SD).
(D–F) Seedlings were treated for 24 hr with mock, 100 nM BL, 5 μM IAA, or 5 μM IAA + 100 nM BL.
(D) Root tip morphology of Col after the treatment. Arrowheads indicate QC cell layer. The scale bar represents 50 μm.
(E) Average of cortex cell length and width of cell numbers 1–15 counted from cortex-endodermis initials (CEIs) (n > 12, mean ± SD).
(F) Average measurement of cortex cell length along the root axis, counted from CEIs (n > 12). Arrows indicate initiation of the elongation zone.
At the cellular level, auxin reduced cell elongation and increased lateral cell expansion in the meristem zone and radial dimension of root tip, in contrast to BR (Figures 7D and 7E). Furthermore, auxin dramatically inhibited cell elongation in the elongation zone. The roots treated with both BL and auxin showed double QC and early initiation of elongation in the transition zone, similar to BL-treated roots, but failed to elongate further in the elongation zone compared to BL-treated roots (Figure 7F).
Discussion
Our study reveals an important role for BR as a major spatiotemporal signal that, through patterning the transcriptional activity of BZR1, regulates the dynamics of stem cell maintenance and differentiation in Arabidopsis roots. We further show that, in contrast to their synergistic interaction in regulating shoot growth, BR and auxin distribute with opposite gradient and interact antagonistically in controlling gene expression, stem cell maintenance, and cell elongation in root tips. We propose that the opposite gradients of these antagonistic signals determine the developmental gradient of stem cells in plant root tips.
Previous studies showed that at different concentrations, BR can have inhibitory and promotive effects on root growth [15, 17]. Here, our experiments show that such dose-dependent opposite effects of exogenous BR are due to requirement of different BR levels in different developmental zones. Under normal conditions, a high level of BR signaling in the transition-elongation zone is required to promote cell elongation, and a low BR level in the QC and meristem area is required to maintain stem cell and meristem size. Thus, the inhibitory and promotive effects of endogenous BR are controlled spatiotemporally to maximize root growth. Treatment with a high concentration of exogenous BR accelerates the transition from meristem cell division to cell elongation, causing an initial increase of root growth rate followed by decreased growth rate due to consequent reduction of meristem size. To avoid the inhibitory effects on QC function and meristem size while promoting cell elongation in the elongation zone, endogenous BR signaling is apparently maintained at a low level in the QC and meristem zone but at a high level in the transition-elongation zone, as indicated by the pattern of nuclear BZR1 and BZR2/BES1. Expression of active bzr1-1D in specific cell types confirmed the requirement of high BR/BZR1 activity in the epidermis of the elongation zone and low BR/BZR1 activity in the QC for normal functions of these cells, namely elongation and quiescence, respectively.
Our genomic analyses provide strong evidence supporting a major role of patterned BZR1 activity in spatial patterning of root gene expression. Comparison among BR-responsive transcriptomes, genome-wide BZR1 targets, and the RootMap transcriptomes revealed a high proportion of the developmental zone-specific genes being BR-regulated and BZR1 targets. Furthermore, BZR1 mainly promotes expression of target genes in the elongation zone but represses target genes in the QC and surrounding cells (Figure 6D). The high level of nuclear BZR1 in the transition-elongation zone correlates with expression of BR-induced genes, many of which encode cell-wall proteins that potentially mediate cell elongation. The low levels of nuclear BZR1 in cells of the QC and meristem zone correlate with expression of a large number of BR-repressed genes, which include genes involved in ribosomal biogenesis and several key stem cell regulators in the QC. These results suggest that BZR1 contributes to the patterning of gene expression by having not only different nuclear levels but also different target gene specificity and transcriptional activity in different cells.
The patterning of nuclear BZR1 depends on BR and involves localized BR catabolism. In BR-deficient plants, the pattern of nuclear BZR1 is abolished but can be re-established from a low concentration of exogenous BR, presumably by BR transport or catabolism, and/or different BR sensitivity. A role of BR catabolism in patterning BZR1 is confirmed by the increased nuclear BZR1 levels in the QC and stem cell regions of the bas1;sob7 double mutant, which is defective in two BR-inactivating enzymes expressed in the root tip region. A recent study showed that another BR-inactivating enzyme, BAT1, is also expressed in the root tip region [45]. While BR synthesis and catabolism are required for normal BZR1 patterning, differential BR sensitivity may also contribute to the BZR1 patterning, as BR treatment and the bzr1-1D mutation reduced, but did not completely abolish, the BZR1 gradient. It is worth noting that localized BR catabolism also inactivates BZR1 in the shoot apical meristem to facilitate stem cell arrest and/or quiescence during organ boundary formation [46, 47].
The patterning of BR/BZR1 appears to also involve auxin, which has been considered the master patterning signal for root development, with a maximum in the root stem cell niche [4, 44, 48]. The BZR1 pattern is overall opposite to that known for auxin and thus can potentially be established by an inhibitory effect of auxin on BR level or signaling. Indeed, auxin treatment increased cytoplasmic localization of BZR1 in the elongation zone, and inhibiting auxin biosynthesis increased nuclear BZR1 in the root tip region, where the endogenous auxin level is at a maximum. Auxin increases the expression levels of several BR catabolic enzymes, including BAS1, SOB7, BAT1, and BEN1 (Table S4; [45]). These observations suggest that the auxin gradient contributes to the complementary pattern of nuclear BZR1 in root tip, at least in part by promoting local BR catabolism. In contrast, BR has been reported to promote auxin transport [21]. Such auxin inhibition of BR and BR promotion of auxin transport could potentially create a feedback loop that helps maintain their balance.
Recent studies showed that the TDIF peptide-activated TDR receptor kinase directly phosphorylates and activates BIN2, leading to phosphorylation and cytoplasmic localization of BZR1 in the procambial cells [35]. However, in contrast to BR, treatment with TDIF did not affect BZR1 pattern in the root tip (Figure S7), consistent with specific function of TDIF-TDR in xylem development. Similarly, cytokinin and ethylene, which both regulate root development through regulating auxin, showed no obvious effect on BZR1 patterning (Figure S7).
Consistent with their opposite gradient patterns and opposite effects on BZR1 nuclear localization, BR and auxin show antagonistic interactions at the transcriptomic level, as well as at cellular and genetic levels, in roots. Both BR and auxin show strong preference of gene activation and repression in different developmental zones that correlate with their endogenous distribution, suggesting major contributions of these hormones to the expression of developmental zone-specific genes. The genes that are oppositely coregulated by BR and auxin include many BZR1 direct target genes, particularly in the QC and transition-elongation zones. In particular, BR/BZR repression of the QC-specific Myb factor BRAVO was recently reported to mediate BR regulation of stem cell quiescence [16], and we found BRAVO expression requires auxin. BR repression of three of the four PLT factors (PLT1, PLT2, PLT4), whose expression requires auxin, is likely to contribute to BR-auxin antagonism in regulating meristem size, as the gradient shape of PLTs determines the boundaries between quiescent and dividing cells and between meristem and elongation zones [44].
The antagonism between BR and auxin on root cell elongation, in contrast to their synergism in shoot cell elongation, reflects a consistent function of BR in promoting cell elongation but opposite effects of auxin in promoting shoot cell elongation and inhibiting root cell elongation, which is apparently necessitated by the opposite gravitropic and phototropic responses of the two organs. Recent studies suggest that distinct mechanisms are involved in auxin regulation of developmental zonation and rapid cell elongation responses in gravitropism [44]. Our study suggests that BZR1 nuclear localization is regulated effectively and rapidly by BR but is modulated slowly and perhaps indirectly by auxin. Thus, BZR1 appears to mediate BR-auxin coregulation of developmental zonation, similar to the role proposed for PLTs [44].
The importance of BR-auxin antagonism in root growth is supported by the genetic and physiological interactions between the two hormones. Previous studies of bin2 [26, 27] and iaa7 mutant [28] and our analyses of additional BR mutants showed consistently opposite effects of genetic defects in each pathway on the sensitivities of root to the two hormones. Furthermore, BR-auxin co-treatment alleviated each other's effects on cell elongation and root growth, indicating that the observed root inhibition by high levels of either BR or auxin alone is partly due to an imbalance between these two hormones.
Together, the results support a model that BR and auxin are distributed with opposite patterns and have antagonistic effects on stem cell maintenance and cell elongation in root tip, at least partly by patterning BZR1. Their opposite patterns of graded distribution enhance their opposing cellular activities in restricted developmental zones, and the balance between BR and auxin determines the balance between stem cell maintenance and differentiation and hence root growth rate. Other hormonal and environmental signals may alter auxin, BR, or both to influence root growth rate [6, 38, 49]. Maintaining the BR-auxin balance will likely involve spatiotemporal interactions between the two hormonal pathways at various levels, such as auxin regulation of BR metabolism [25, 50], BR regulation of auxin transport [21], BZR1-mediated transcriptional regulation of auxin catabolic enzymes, and signaling components (Table S5), and possibly direct interaction between signaling components [24, 29, 30]. Clearly, a complete understanding of how the BR-auxin circuitry regulates plant development will require future studies carried out in specific developmental contexts.
Experimental Procedures
Plant Growth and Root Imaging
All wild-type, various mutants, and transgenic lines are in the Arabidopsis thaliana Columbia (Col-0) background (for details of plant lines and plasmid construction, see Supplemental Experimental Procedures). Seeds were surface sterilized and plated on 0.5× MS media containing 1% sucrose, 0.8% Phytoblend agar,and indicated hormones or mock. Plates were placed vertically under constant light. Root lengths were measured from images using ImageJ. For live-imaging analysis, root growth rates were measured every 15 min for 3 days (see Supplemental Experimental Procedures for more details).
Confocal Microscopy and Quantification of Fluorescence Signal
Laser scanning confocal microscopy was performed by using a Leica SP5 system after propidium iodide staining. At least five roots were analyzed for each treatment in each experiment, and each experiment was repeated independently at least three times. The same laser settings were used within the same experiment. To determine N/C ratios of BZR1-YFP, BES1-GFP, and bzr1-1D-CFP, we crossed the transgenic lines with plants expressing a nuclear marker 35S:H2B-RFP, and serial optical sections were obtained from roots of F1 seedlings. Meristem zone was between QC and the transition zone (cells in square shape). Elongation zone was between the transition zone and the first cell with root hair bulge.
Gene Expression Analysis
Total RNA was extracted from root tips of 7-day-old seedlings treated with 100 nM BL, 5 μM IAA, 100 nM BL + 5 μM IAA, or ethanol (mock) solution for 4 hr using the Spectrum Plant Total RNA kit (Sigma). For qRT-PCR, the total RNA was used to prepare cDNA using RevertAid reverse transcriptase (Fermentas). qRT-PCR was performed on LightCycler 480 (Roche) using a SYBR Green reagent (Bioline) (see Supplemental Experimental Procedures for qRT-PCR primers). For RNA-seq, libraries were constructed using the TruSeq RNA Sample Preparation kit (Illumina) according to manufacturer's instructions. Reads were aligned to the Arabidopsis genome, TAIR10. Read counts for each gene were quantified, and differential expression was determined using fold change > 1.4 (equivalent to log2 fold change > 0.5) and p < 0.01 cutoffs (see Supplemental Experimental Procedures for detailed analysis).
Supplementary Material
Highlights.
Spatially patterned BR signaling orchestrates stem cell dynamics and root growth
Root tip has a gradient of nuclear BZR1 with opposite transcriptional activities
BR and auxin have opposite patterns and effects on cell elongation in root tip
A BR-auxin balance is required for balanced stem cell dynamics in root meristem
In Brief.
The study by Chaiwanon and Wang provides evidence for a brassinosteroid (BR) gradient contributing to the developmental gradient of root meristem in Arabidopsis. A balance between the oppositely patterned, antagonistic actions of BR and auxin maintains the spatiotemporal balance between stem cell maintenance and differentiation in root meristems.
Acknowledgments
We thank E. Oh for making the pTOPO-bzr1-1D-YFP construct; Y. Sun for making pBES1:BES1-GFP lines; J. Dinneny for the SCR and WER promoters, the erGFP constructs, and the pGoldenGate-SE7 vector; H. Cartwright for assistance with confocal image quantification; Y. Geng for assistance with live imaging; M. Neff for the bas1-2/sob7-1 seeds; A. Caño-Delgado for the pBRAVO:GFP seeds; B. Scheres for the pPLT1:PLT1-YFP and pPLT2:PLT2-YFP seeds; J. Haseloff for the 35S:H2B-RFP seeds. We thank D. Bergmann, D. Ehrhardt, J. Dinneny, and S. Long for comments on the manuscript. This study was supported by a grant from the NIH (R01GM066258 to Z.-Y.W.).
Footnotes
Accession Numbers: The GEO accession number for the RNA-seq data reported in this paper is GSE52966.
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures, seven figures, five tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2015.02.046.
References
- 1.Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Beemster GT, Baskin TI. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998;116:1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubeda-Tomás S, Beemster GTS, Bennett MJ. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci. 2012;17:326–331. doi: 10.1016/j.tplants.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- 6.Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 7.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 8.Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster GTS, Bhalerao R, Bennett MJ. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- 9.Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI. Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J. 2007;50:514–528. doi: 10.1111/j.1365-313X.2007.03068.x. [DOI] [PubMed] [Google Scholar]
- 10.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Bargmann BOR, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, Birnbaum KD. A map of cell type-specific auxin responses. Mol Syst Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clouse SD, Sasse JM. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 13.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müssig C, Shin GH, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133:1261–1271. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- 16.Vilarrasa-Blasi J, González-García MP, Frigola D, Fàbregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell. 2014;30:36–47. doi: 10.1016/j.devcel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138:839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, De Veylder L. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342:860–863. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- 19.Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- 20.Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA. 2009;106:13630–13635. doi: 10.1073/pnas.0906416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134:1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- 26.Pérez-Pérez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- 27.Maharjan PM, Schulz B, Choe S. BIN2/DWF12 antagonistically transduces brassinosteroid and auxin signals in the roots of Arabidopsis. J Plant Biol. 2011;54:126–134. [Google Scholar]
- 28.Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 2006;45:193–205. doi: 10.1111/j.1365-313X.2005.02582.x. [DOI] [PubMed] [Google Scholar]
- 29.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. [DOI] [PubMed] [Google Scholar]
- 31.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 32.Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 35.Kondo Y, Ito T, Nakagami H, Hirakawa Y, Saito M, Tamaki T, Shirasu K, Fukuda H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat Commun. 2014;5:3504. doi: 10.1038/ncomms4504. [DOI] [PubMed] [Google Scholar]
- 36.Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 2005;42:23–34. doi: 10.1111/j.1365-313X.2005.02358.x. [DOI] [PubMed] [Google Scholar]
- 37.Bennett T, Scheres B. Root development-two meristems for the price of one? Curr Top Dev Biol. 2010;91:67–102. doi: 10.1016/S0070-2153(10)91003-X. [DOI] [PubMed] [Google Scholar]
- 38.Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 39.He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 42.Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell. 2005;17:2340–2354. doi: 10.1105/tpc.105.033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffith ME, Mayer U, Capron A, Ngo QA, Surendrarao A, McClinton R, Jürgens G, Sundaresan V. The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell. 2007;19:2246–2263. doi: 10.1105/tpc.106.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515:125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi S, Cho YH, Kim K, Matsui M, Son SH, Kim SK, Fujioka S, Hwang I. BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J. 2013;73:380–391. doi: 10.1111/tpj.12036. [DOI] [PubMed] [Google Scholar]
- 46.Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA. 2012;109:21146–21151. doi: 10.1073/pnas.1210789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 49.Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, et al. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 2011;66:564–578. doi: 10.1111/j.1365-313X.2011.04513.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


