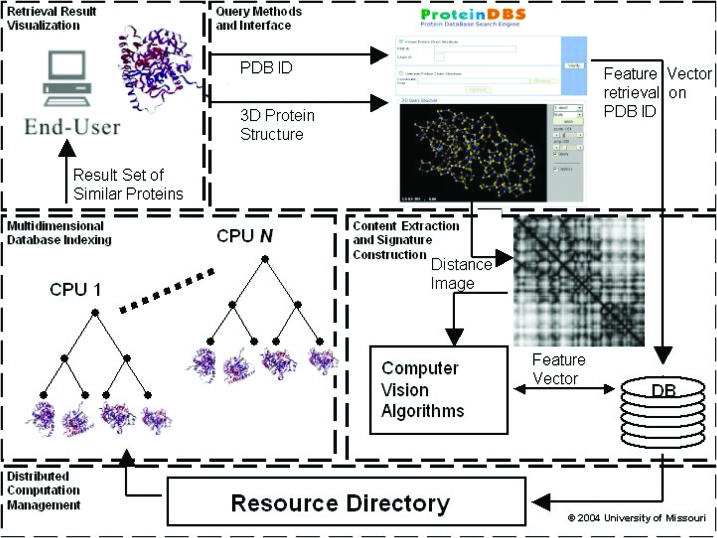
Figure 1.
ProteinDBS system diagram. An end-user submits a protein structure encoded in the PDB format or the PDB ID of a protein structure. Given a protein structure PDB file, the interface converts the 3D coordinate structure into a 2D distance matrix image. The application of various computer visualization algorithms produces a protein signature that is used as a query against the database. The protein structure, either computed or retrieved from a relational Database Management System (DBMS) given the PDB ID, is used to determine a similarity-ranked retrieval set from the distributed EBS k-d tree indices. The most similar proteins are then displayed back to end-user.