Fig 5. Impact of low-density microarray data on the determination of the nearest neighbors.
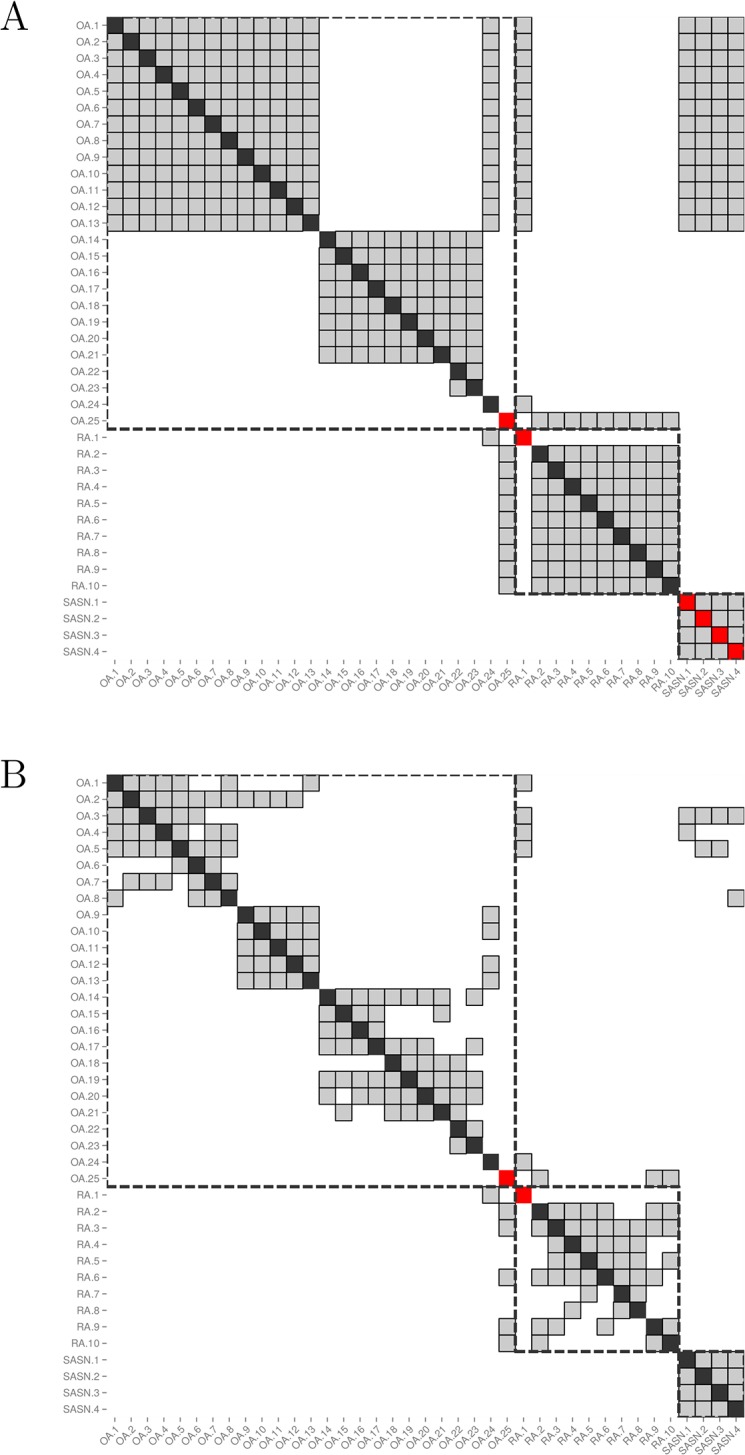
Both matrices show the nearest neighbors of each biopsy sample from the cohort of patients with a known diagnosis (n = 39). Each sample is represented by a column, and its nearest neighbors are greyed out in that column. The cell on the diagonal is red if the sample is misclassified and black otherwise. Samples from patients with the same diagnosis are surrounded by a dashed square. (A) Nearest neighbors are determined using clinical data only (ρ = 1). More than 5 nearest neighbors are displayed for each sample due to the presence of ties. (B) Nearest neighbors are determined using a combination of clinical and low-density array data (ρ = 0.5), resulting in a correct classification of the last 4 SA samples thanks to good tie-breaking.
