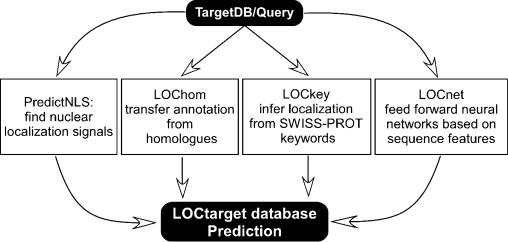
Figure 1.
The LOCtarget system. From the query amino acid sequence, the three-state secondary structure and solvent accessible surface residues of the protein are predicted using PROFphd (29; B. Rost, manuscript submitted). LOCtarget uses four different methods to annotate sub-cellular localization. (i) PredictNLS: the amino acid sequence is scanned for nuclear localization signals. (ii) LOChom: the sequence is first aligned through PSI-BLAST profiles to a database with experimental annotations about localization. If any sequence homologues are discovered, sub-cellular localization annotation is transferred from the homologue. (iii) LOCkey: the SWISS-PROT database contains functional information for proteins in the form of keywords. LOCkey infers sub-cellular localization based on keyword entries. The above three programs are based solely on the amino acid sequence of the protein and do not use any structural information. (iv) LOCnet: sub-cellular localization is predicted by a system of neural networks trained on a number of global features such as amino acid composition, predicted secondary structure composition and composition of predicted surface accessible residues. The final localization annotation in the LOCtarget database is taken from the most reliable prediction amongst the four individual methods.