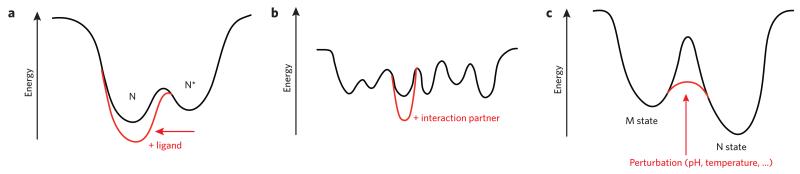
Figure 1. Schematic two-dimensional functional energy landscapes.
(a) Many proteins sample a near-native state (N*) to open a binding site and interact productively with a ligand. Formation of the complex with ligand stabilizes the protein in its native state (N). (b) IDPs have sequences that disfavor a unique folded state and instead lead to many conformational possibilities of nearly equivalent energy. Interaction(s) with partners leads to stabilization of the state(s) that have the capacity to bind the partner with the highest affinity, thus shifting the energy landscape in the presence of partner(s) to one or only a few more stable states. (c) The magnitude of barriers on energy landscapes has a profound impact on protein function. A state that is not the thermodynamically most stable state (here M) may be long lived because it is separated from the most thermodynamically stable state (N) by high-energy barriers. The barriers may be reduced in amplitude by various triggers, whether environmental or protein-protein interactions.