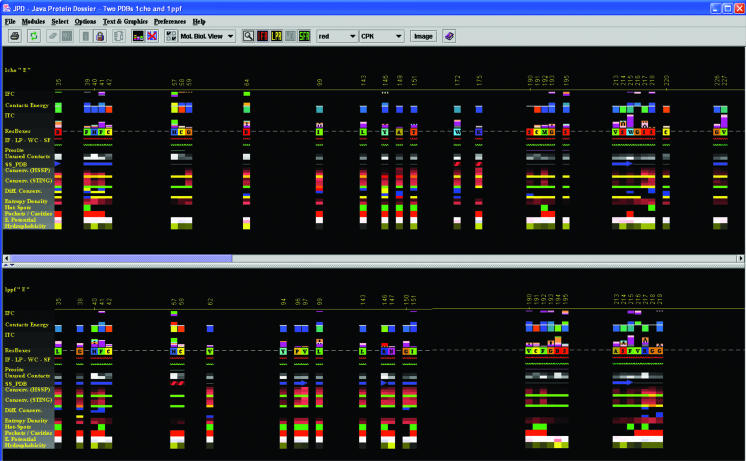
Figure 2.
Structural alignment in JPD. This is one of the key features that JPD offers to users—the possibility to structurally align two proteins and consequently display ‘aligned’ structural parameters. In this case, we used two serino proteases: Chymotrypsin in complex with turkey ovomucoid third domain (1cho.pdb) and elastase in complex with the same inhibitor (1ppf.pdb). As a result of the alignment, Gold Sting shows, in the Chime window, two aligned structures and, in the Gold Sting sequence window, the sequences of two chains, aligned according to the structural alignment. This particular picture was obtained after clicking on the IFR icon in the JPD speed icons area. The displayed pattern shows only the residues at the interface area of the two aligned chains. After the user selects to view only the IFR region, JPD continues to preserve the alignment mode, so that the user can inspect the spatial position of these important ‘interfacing’ residues (of course in comparison with the homologous and aligned structures). This clearly offers a rapid insight into the mechanism of the specificity for certain substrates/inhibitors. In this specific case, we see two different enzymes binding to the same inhibitor. Both enzymes demonstrate a similar pattern of IFRs, yet variations between the two ensembles of IFRs should suggest the key elements responsible for the successful binding of the inhibitor to both enzyme chains.