Figure 4.
RNF4 Is a Regulator of the FA Pathway and Polyubiquitylates the SUMOylated ID Complex
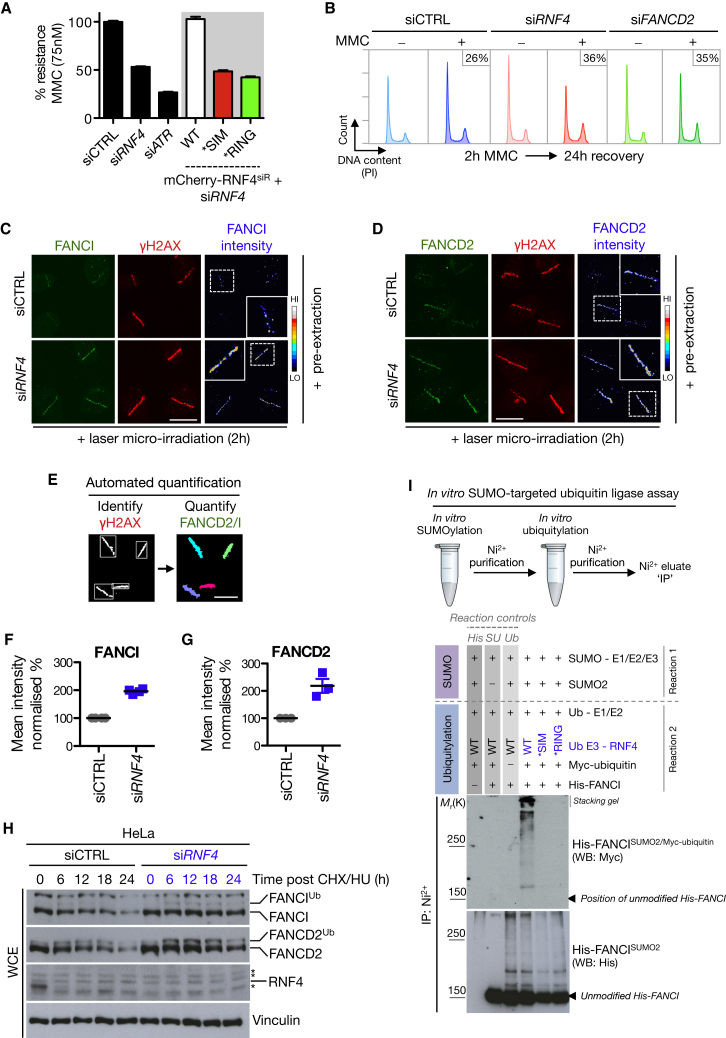
(A) Cellular fitness of U2OS cells transfected with control (CTRL), ATR, or RNF4 siRNA was assessed using the multicolor competition assay (MCA) (Smogorzewska et al., 2007). Stable U2OS/mCherry-RNF4 siRNA-resistant (siR) cell lines were used for RNF4 allele complementation analysis. Data represent mean ± SEM from three independent experiments
(B) HeLa cells were transfected with the indicated siRNAs, exposed to a pulse of MMC (50 ng/ml) for 2 hr, and then allowed to recover for 24 hr before fixation and analysis by flow cytometry. The proportion of cells in G2/M phase is indicated.
(C) U2OS cells transfected with control (CTRL) or RNF4 siRNA were subjected to laser microirradiation, pre-extracted and fixed 2 hr later, and immunostained with FANCDI and γH2AX antibodies. Scale bar represents 10 μm.
(D) Same as (C) except with FANCD2 antibody.
(E) QIBC strategy to analyze data generated from laser microirradiation experiments in an automated unbiased manner.
(F) Quantification of normalized mean FANCI intensities at sites of laser microirradiation. Each data point represents the quantification of 75–150 cells from three independent experiments.
(G) Same as (F) but for FANCD2.
(H) HeLa cells transfected with control (CTRL) or RNF4 siRNA were treated with cycloheximide (CHX) and HU for the indicated times. Protein extracts were analyzed by immunoblotting with indicated antibodies. ∗, denotes cross-reactive bands.
(I) His-FANCI was SUMOylated in vitro as in Figure 2D, purified on Ni2+ agarose and then subjected to in vitro ubiquitylation by RNF4. The Ni2+ beads were then washed extensively and analyzed by immunoblotting with indicated antibodies.
See also Figure S4.