Figure 1.
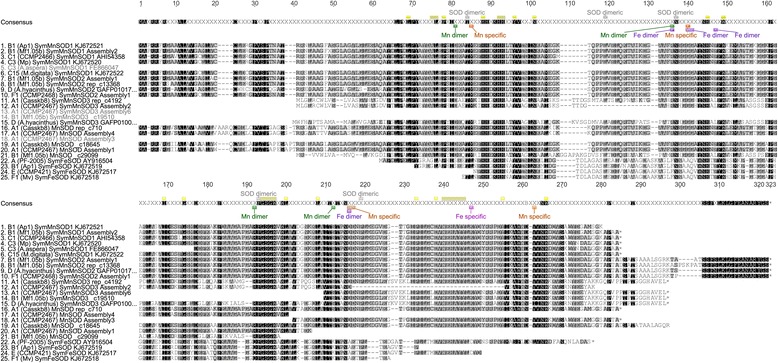
Alignment of MnSOD (sequences 1–21) and FeSOD (22–25) protein isoforms from different Symbiodinium ITS2 types. Residues used for identification of metalloform are highlighted on the consensus sequence. Conserved SOD residues (yellow), SOD dimer-specific Thr84, Leu119, Asn137, Phe193, and Pro219 (grey), MnSOD specific Met85, Gly140, Gln216, Asp217, Val263 (orange), FeSOD specific Asp247 (magenta), manganese dimer-specific Asp81, Arg136, Arg192, and Ser212 (green), and iron dimer-specific residues Phe136, Ala140, Gln141, Phe147, Ala216 (purple) have all been highlighted. Annotations were deduced from alignments with 17 characterized iron and manganese SOD sequences [55]. Ends of ORFs are indicated by asterisks. Shading indicates site-specific similarity over all sequences 100% (black), 80-100% (dark grey), 60-80% (light grey), and less than 60% (white), based on the Blosum62 score matrix with a threshold of 1. Sequence IDs contain ITS2 type, strain designation or source of isolation (in brackets) and GenBank accession number or contig/assembly designation (Additional file 11).
