Fig 1. Phylogenetic alignments of LCLs to EBV-1 and EBV-2.
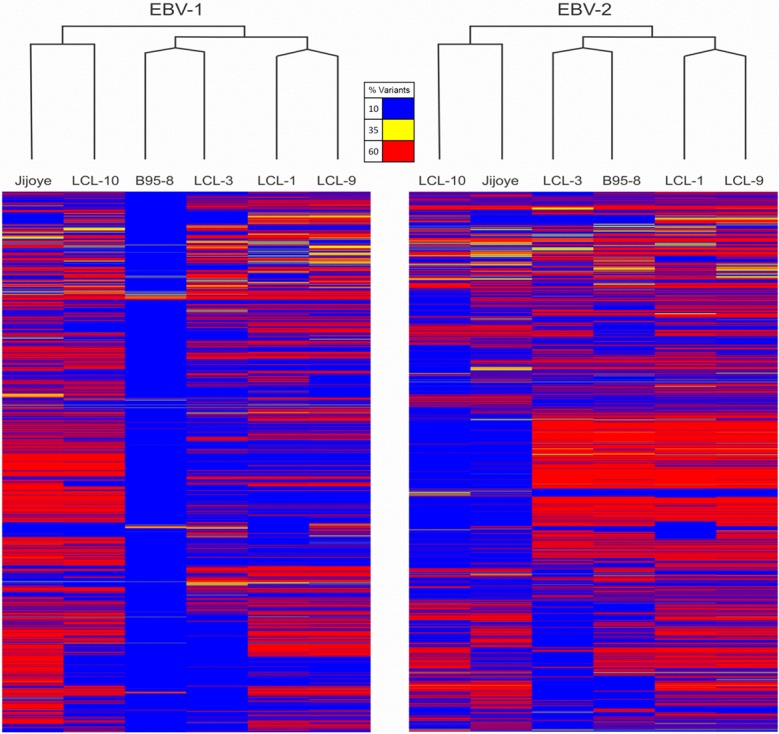
The figure shows the alignment of the LCLs, B95.8 cell line and Jijoye cell line controls against EBV-1 and EBV-2. It is observed that B95.8 aligned closely to EBV-1, followed by LCL-3, LCL1 and LCL9, while LCL10 and Jijoye were distant from EBV-1. B95.8 is clustered with LCL3, LCLI and LCL9 are clustered together, while LCL10 and Jijoye are clustered together when compared to EBV-1. There were relatively less base changes in the middle for most of the LCLs except LCL10. Similar trend was observed on comparison with EBV-2, however, LCL10 was much closer to EBV-2 reference than Jijoye and the clustering was maintained as with EBV-1 reference. The bases were more conserved in the middle region of the genome than in the N and C terminus for LCL10 and Jijoye, for B95.8, LCL1, LCL3, and LCL9 the base changes were spread all over the genome with majority of the changes in the middle. Additionally, LCL3 was more conserved at the C-terminal end of the genome, as observed with EBV-1.
