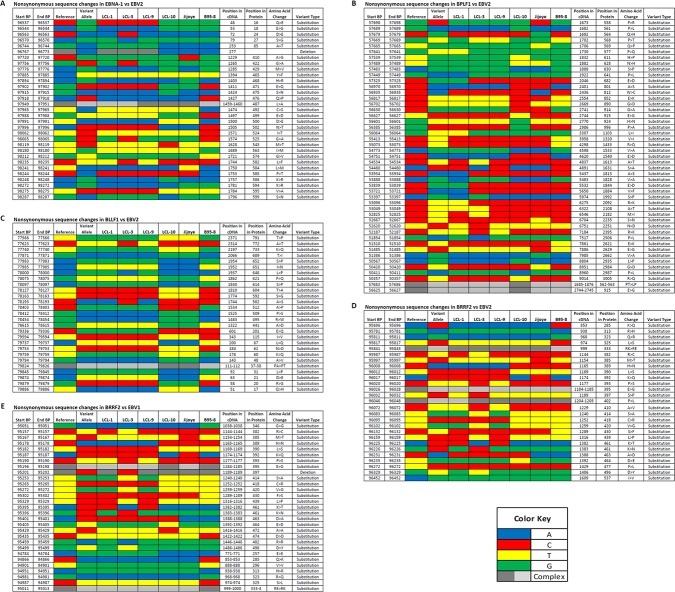
Fig 5. Non-synonymous sequence changes.
(A) EBNA1 compared to EBV-2. Using heat maps in our analysis we observed that most EBNA1 mutations were substitutions with a single deletion (96733), and that the changes were similar in all LCLs except in a few cases where there were changes in LCL3 (97756) and in LCL10 (98119, 98248, and 98287) distinct from the other LCLs. (B) BPLF1 compared to EBV-2. When aligned to EBV-2 all the mutations observed in BPLF1 were substitutions. All the substitutions were generally similar in the LCLs, with some changes seen in LCL3 alone (54534, 53397, 52825, and 51854), LCL10 alone (57689, 57539, 56627, 56385, 50567, 50411, 50357, 57686, and 56 627). (C) BLLF1 compared to EBV-2. All the BLLF1 mutations observed were substitutions and they were mostly shared by all LCLs. LCL10 and Jijoye were perfect match with the reference, the rest varied with the reference genome at different sites. LCL1, LCL3, and LCL9 all had similar changes compared to reference except LCL3 at 77623, 78193, and 78000 that were similar to reference. (D) BRRF2 compared to EBV-2. We observed that when compared to EBV-2 reference, all the BRRF2 mutations were substitutions, which were similar in most LCLs except for a couple specific for LCL3 (95987 and 96329), and LCL10 (96272). (E) BRRF2 compared to EBV-1. When BRRF2 was compared to EBV-1, most of the changes were substitutions, but we had a deletion too that was detected in all samples. Again LCL3 had specific mutations (95157, 95396, 95459, and 95499) and LCL10 (95429). It is to be noted that only LCL3 had no deletion at 95201.