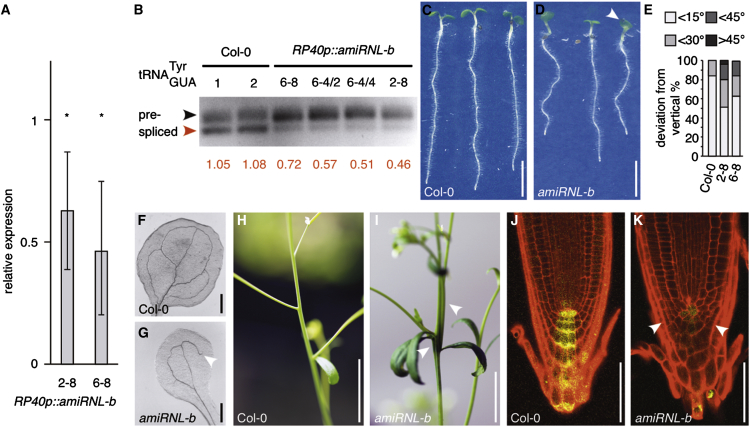
Figure 5.
Analysis of AtRNL RNA Ligase Knockdown Lines
(A) qPCR displaying AtRNL transcript levels in RP40p:amiRNL-b lines 2–8 and 6–8 at 6 DAG (Col-0 = 1). Results from three biological repeats with three technical replicates are shown. SDs are indicated (∗Student’s two-tailed t test, p < 0.01).
(B) Quantification of levels in Col-0 and RP40p::amiRNL-b silencer lines by PCR. Upper band corresponds to non-spliced precursor (black arrowhead); lower band corresponds to processed (red arrowhead). Ratios of spliced and precursor band intensities are displayed below.
(C and D) Col-0 (C) and RP40p::amiRNL-b (D) seedlings at 6 DAG. White arrowhead in (D) points to fused cotyledons.
(E) Orientation of primary root growth of Col-0 and RP40p::amiRNL-b lines 2–8 and 6–8 seedlings at 8 DAG. A total of 40–50 seedlings was analyzed for each genotype and plotted as percentage of seedlings displaying <15°, <30°, <45°, and >45° deviation from the vertical growth axes.
(F and G) Cotyledon vasculature in Col-0 (F) and RP40p::amiRNL-b (G). White arrowhead indicates vasculature discontinuity not observed in wild-type.
(H and I) Detail of inflorescence axes of flowering Col-0 (H) and RP40p::amiRNL-b (I) at 28 DAG. White arrowheads indicate fused, fasciated stem.
(J and K) Comparison of wild-type (J) and RP40p::amiRNL-b (K) primary root meristems expressing auxin-responsive DR5rev::GFP (yellow). Roots were stained with PI (red) to visualize cell boundaries. White arrowheads point to alterations in root meristem patterning.
Size bars represent 10 mm (C, D, H, and I), 1 mm (F and G), and 50 μm (J and K). See also Figures S5 and S6.