Extended Data Figure 4. Transgenic P. falciparum expressing PfKelch13C580Y mutation in two different strains of P. falciparum using distinct approaches and markers.
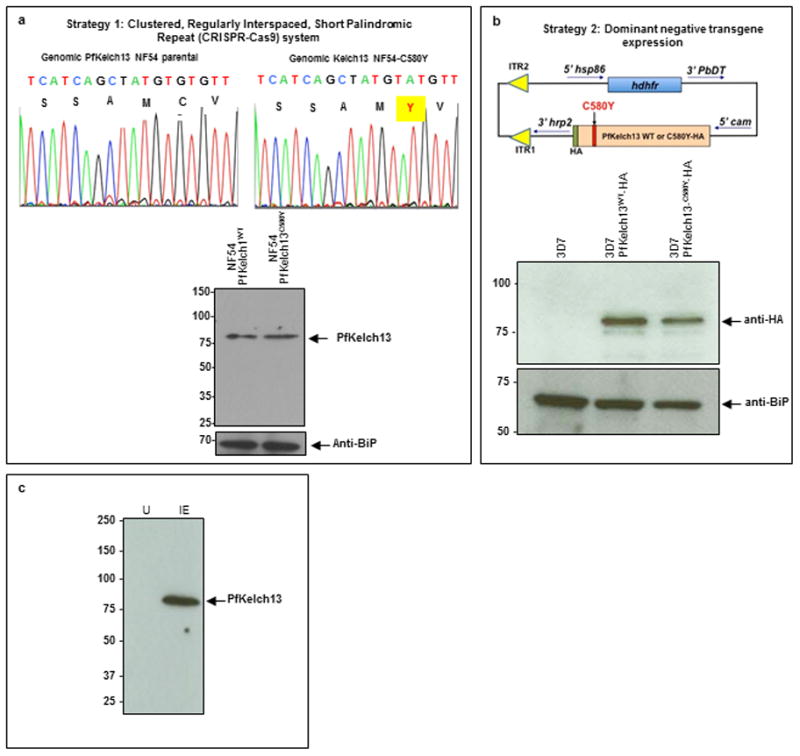
a, Strategy 1: CRISPR-Cas9 used to introduce a single point mutation in the P. falciparum NF54 genome as described elsewhere20. Both parent and mutant strains were sequenced to ensure that the only change detected was PfKelch13C580Y. Western blots using anti-PfKelch13 antibodies confirm that mutation does not change the level of PfKelch13 protein. b, Strategy 2 expresses a dominant negative PfKelch13C580Y and wild type PfKelch13WT tagged with HA, and driven by the constitutive cam promoter in P. falciparum 3D7. Western blots confirm that anti-HA antibodies detected tagged forms of PfKelch13 in transfected but not non-transfected 3D7. Comparable levels of wild type and mutant transgenes are expressed in resulting two strains of parasites (as shown). In a-b, BiP, an ER marker serves as a parasite loading control in western blots. Molecular weight standards (in kDa) are as indicated. c, Western blot indicate that antibodies to PfKelch13 specifically recognize a ~83-kDa protein in infected (IE) but not uninfected erythrocytes (U). Molecular weight standards (in kDa) are as indicated. In a-c: data are representative of three experimental replicates; raw data are in Supplementary Data 2.
