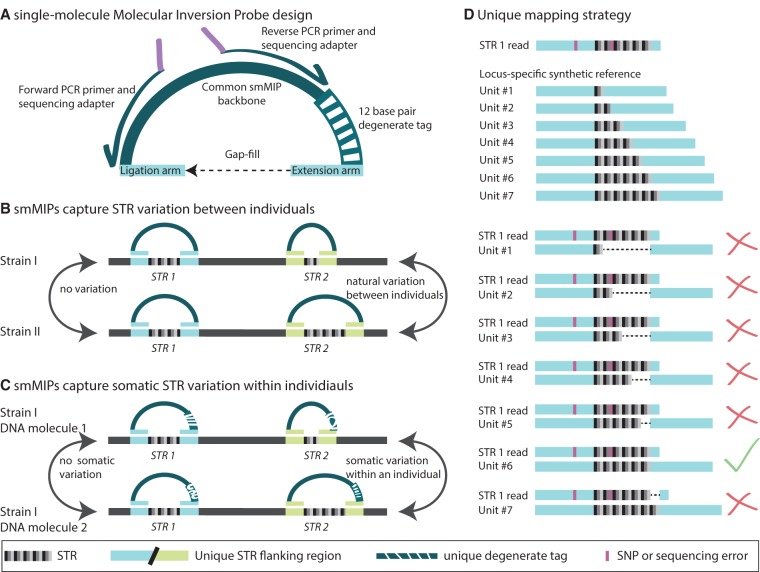
Figure 1.
MIPSTR determines germline and somatic STR variation through targeted capture, sequencing, and a novel mapping strategy. (A) Single-molecule molecular inversion probe (smMIP) with common backbone for PCR primer binding (dark-green; also shown, PCR and sequencing primers with arrows and purple sequencing adapters); 12 base pair degenerate tag (striped, green/white); and targeting arms with locus-specific, STR-flanking sequence (blue). One targeting arm is the primer for polymerase extension (extension arm). Ligation closes the circle at the other targeting arm (ligation arm). (B) Capture across genetically diverse individuals identifies germline STR variation across genetically diverse individuals. (C) MIPSTR distinguishes somatic STR variation from technical error, using many degenerate tags. STR variation within a tag-defined read group (i.e., reads with the same degenerate tag) is considered technical error. STR variation across tag-defined read groups is considered somatic variation. (D) MIPSTR maps reads from a given STR locus (based on targeting arm sequence) to its locus-specific synthetic reference with unit numbers 1–100 (1–7 shown here). The STR 1 read aligns perfectly to locus-specific synthetic reference Unit #6 (green check mark); all other alignments show gaps (dashed line, red X). SNVs (in pink), even if occurring in the STR sequence, do not affect mapping or STR unit number genotype calls.