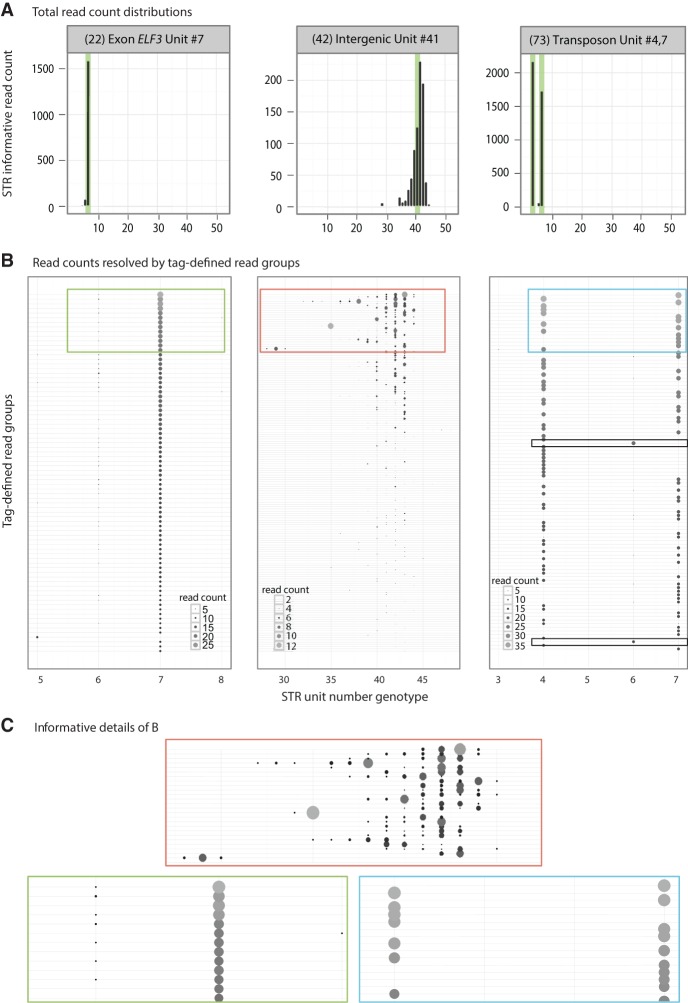
Figure 3.
MIPSTR distinguished technical error from somatic variation. (A) Three histograms from Figure 2 with total read counts. (Left) The known ELF3 STR unit number is clearly supported by the modal unit number. (Middle) This intergenic STR showed great variation in STR unit number; the mode did not support the known STR unit number. (Right) This STR resides in two copies in two different genomic locations (transposons). Both known alleles were identified, yet total read counts alone cannot distinguish genomic duplicates from technical or somatic error. (B) Reads are separated into tag-defined read groups with dot sizes and shading representing read count (different scales for each locus, see inset). Colored boxes are shown in detail in C. (Left) All tag-defined read groups with one exception supported the known STR unit number seven. Most tag-defined read groups showed low levels of technical error, primarily reads with unit number six (-1), but also five and eight. (Middle) Separating reads into tag-defined read groups illustrates the extremely high technical error for this STR. The mode of a tag-defined read group was often supported by <50% of total reads. Some tag-defined read groups contained as many as six different STR genotypes. We exclude such loci from the analysis of somatic STR variation. (Right) As expected for a duplicate STR or a heterozygote, approximately half of the tag-defined read groups support each of the known STR genotypes with very little technical error. We also observed evidence of a somatic STR allele with unit number six, which was supported by two tag-defined read groups (boxed, black outline). Note the absence of either of the known STR alleles for these tag-defined read groups. This STR genotype is also visible in the total read count histogram (A, right), where it would be interpreted as a technical error by other methods. (C) Detailed views of plots in B; outline color corresponds to respective plot.