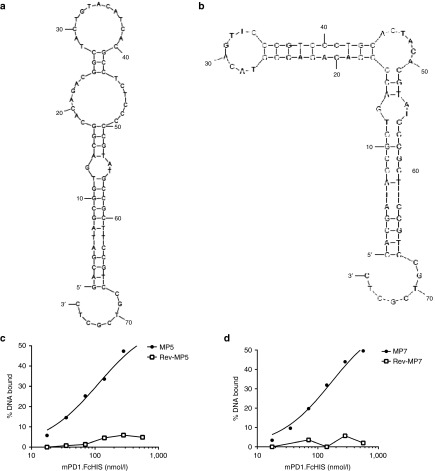
Figure 1.
Secondary structure and binding affinities of highly enriched anti-PD-1 aptamer sequences MP5 and MP7. Mfold40 secondary structure predictions of the top two enriched aptamer sequences MP5 (a) and MP7 (b) identified by next-generation sequencing after five rounds of SELEX toward mPD-1.FcHIS. (c,d) Concentration-dependent binding of 32P-labeled aptamers MP5 (c) and MP7 (d) to mPD-1.FcHIS captured in a nitrocellulose filter binding assay. Filled symbols represent the % DNA bound of the full-length aptamer at each mPD-1.FcHIS concentration while empty symbols represent the % DNA Bound of the respective reverse complement of each aptamer used as a negative binding control.