Figure 3.
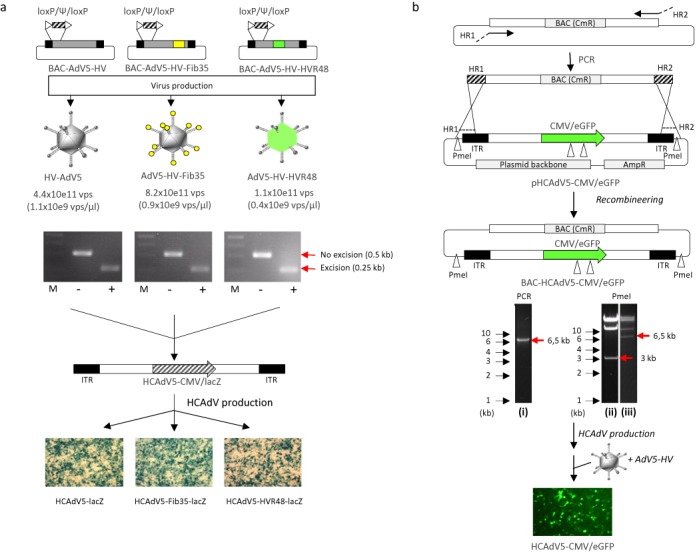
Exploring the BAC platform for cloning and production of HCAdV. (a) Generation of capsid-modified HV with a floxed packaging signal using the BAC platform. A floxed packaging signal (loxP/Ψ/loxP) was inserted into unmodified and capsid-modified BACs (BAC-AdV5, BAC-AdV5-Fib35, BAC-AdV5-HVR48) (see also Figure 2) by recombineering resulting into BAC-AdV5-HV, BAC-AdV5-HV-Fib35, and BAC-AdV5-HV-HVR48. Respective helper viruses were produced and excision of the packaging signal was shown by PCR. Agarose gels show PCR products after infection with HVs of cells either lacking Cre recombinase (−) or stably encoding Cre recombinase (+). HVs were used to produce a capsid modified HCAdV (HCAdV5-CMV-lacZ) lacking all viral coding sequences which contains a transgene expressing ß-galactosidase (lacZ) under the control of the cytomegalovirus (CMV) promoter. The lower panel shows ß-galactosidase of cells infected with unmodified (HCAdV-lacZ) and capsid-modified HCAdVs (HCAdV5-Fib35-lacZ and HCAdV5-HVR48-lacZ). ITR: adenoviral inverted terminal repeats, M: DNA ladder. (b) For available plasmids containing adenoviral genomes (e.g. the HCAdV genome), respective BACs can be generated by backbone-exchange, replacing the plasmid backbone with the BAC-specific backbone containing a chloramphenicol resistance marker gene (CmR). Here, the BAC-specific backbone was amplified by PCR utilizing primers to generate regions homologous to the 5′- and 3′-end of adenoviral genome (HR1 and HR2). For recombineering the purified PCR-product was transformed into SW102 bacteria with the plasmid containing the adenoviral genome. Bacteria with BACs containing the adenoviral genome were isolated by positive selection for chloramphenicol resistance. The left agarose gel shows amplification of the PCR-product used for transformation (i). Successful backbone-exchange was verified by PmeI digest (PmeI restriction enzyme recognition sites are indicated by white arrows) of the plasmid (ii) and the generated BAC (iii) containing the genome of a high-capacity AdV genome with an eGFP expression cassette (BAC-HCAdV5-CMV/eGFP). Note that the 700 bp band after PmeI digest is not visible on the agarose gel. Fragments specific for respective backbones are marked with red horizontal arrows. The corresponding HCAdV (HCAdV5-CMV/eGFP) was produced using the helper virus HV-AdV5 and infection of HEK293 cells with the purified HCAdV resulted in eGFP expression.
