Figure 6.
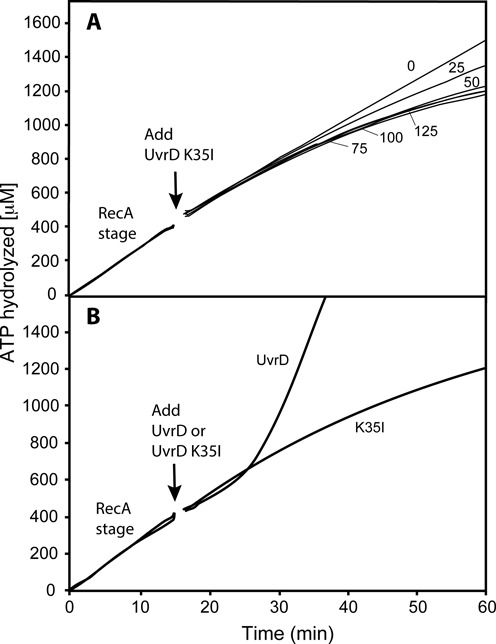
Disassembly of active RecA nucleoprotein filaments in the presence of the ATPase deficient UvrD K35I. (A) Reactions were carried out as described in Materials and Methods, but with the substitution of UvrD K35I protein for WT UvrD. The UvrD variant was added at 15 min (arrow) at the concentration (in nM) shown next to each ATPase curve. The data following this addition have been corrected for a small decline in absorption caused by a dilution effect. The average rate of ATP hydrolysis in the RecA stage is 27 μM/min (yielding an apparent kcat—assuming the 1 μM potential DNA binding sites are all occupied by RecA—of 27 min−1). (B) A comparison between the inhibition of RecA ATPase activity by WT UvrD and UvrD K35I, each added to 50 nM final concentration at 15 min (arrow).
