Figure 8.
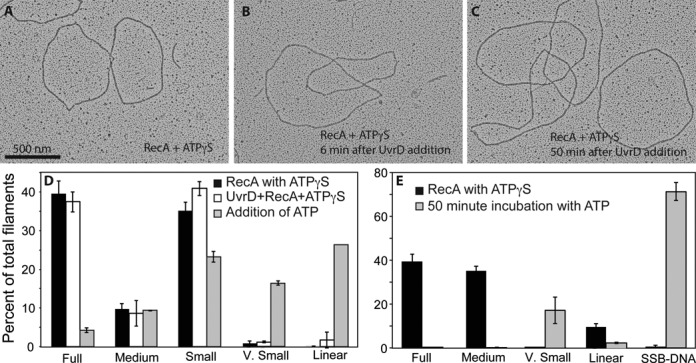
ATP hydrolysis by UvrD is required for dismantling RecA filaments. (A) RecA filaments formed on circular ssDNA after a 10 min incubation with 1 mM ATPγS. Two full filaments and a very small filament, as well as SSB-coated DNA are present in this image. (B) RecA filaments formed as in A), after a six-minute incubation with UvrD. (C) RecA filaments formed as in (A) after 50 min incubation with UvrD. No decrease in filamentation can be observed. (D) If 3 mM ATP is added with the UvrD helicase, significant disassembly of the RecA filaments is seen after 6 min. Molecules are quantified with RecA and ATPγS alone (n = 1027 molecules counted and scored), after addition of UvrD (n = 1187 molecules counted and scored) and after further addition of ATP (n = 1060 molecules counted and scored). (E) Experiment as in panel D, but with less ATPγS (30 μM) added to form the RecA filaments, addition of UvrD, and filaments examined 50 min after further addition of 3 mM ATP. Molecules counted and scored (n) = 573 prior to and (n) = 581 at 50 min after addition of ATP.
